

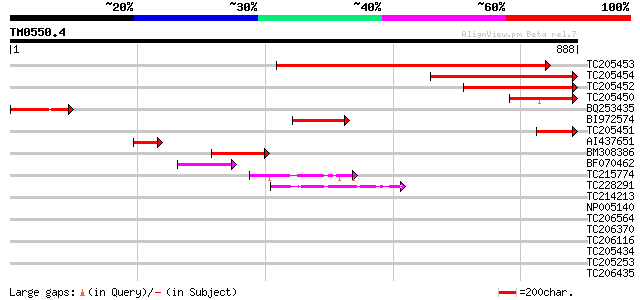
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0550.4
(888 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205453 similar to UP|LON1_ARATH (O64948) Lon protease homolog ... 740 0.0
TC205454 similar to UP|LON1_ARATH (O64948) Lon protease homolog ... 413 e-115
TC205452 similar to UP|LON1_ARATH (O64948) Lon protease homolog ... 310 2e-84
TC205450 similar to UP|Q8GV57 (Q8GV57) Lon protease, partial (12%) 150 2e-36
BQ253435 similar to SP|O04979|LON1_ Lon protease homolog 1 mito... 150 2e-36
BI972574 homologue to SP|P93655|LON2_ Lon protease homolog 2 mi... 121 1e-27
TC205451 similar to UP|Q8GV57 (Q8GV57) Lon protease, partial (7%) 111 2e-24
AI437651 similar to SP|O04979|LON1_ Lon protease homolog 1 mito... 82 1e-15
BM308386 similar to SP|P93648|LON2 Lon protease homolog 2 mitoc... 69 1e-11
BF070462 similar to SP|P93648|LON2 Lon protease homolog 2 mitoc... 54 4e-07
TC215774 similar to UP|Q94ES0 (Q94ES0) AAA-metalloprotease FtsH,... 45 1e-04
TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulato... 43 5e-04
TC214213 homologue to UP|PRSA_BRACM (O23894) 26S protease regula... 41 0.003
NP005140 heat shock protein 40 0.005
TC206564 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 39 0.008
TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 39 0.010
TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 h... 39 0.013
TC205434 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regula... 38 0.022
TC205253 homologue to UP|O99018 (O99018) Chloroplast protease pr... 37 0.038
TC206435 homologue to UP|PRSA_LYCES (P54776) 26S protease regula... 37 0.050
>TC205453 similar to UP|LON1_ARATH (O64948) Lon protease homolog 1,
mitochondrial precursor , partial (48%)
Length = 1290
Score = 740 bits (1910), Expect = 0.0
Identities = 370/430 (86%), Positives = 400/430 (92%)
Frame = +1
Query: 418 LASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDE 477
LASSIAAAL+RKFVRISLGGVKDEADIRGHRRTY+GSMPGRLIDGLKRVAVCNPVMLLDE
Sbjct: 1 LASSIAAALERKFVRISLGGVKDEADIRGHRRTYVGSMPGRLIDGLKRVAVCNPVMLLDE 180
Query: 478 IDKTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLD 537
+DKTGSD+RGDPASALLEVLDPEQNK+FNDHYLNVPFDLSKV+FVATANR Q IPPPL D
Sbjct: 181 VDKTGSDIRGDPASALLEVLDPEQNKSFNDHYLNVPFDLSKVVFVATANRLQPIPPPLRD 360
Query: 538 RMEVIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGMVQLVIQRYTREAGVRN 597
RME+IELPGYTPEEKL IAM+HLIPRVLDQHGLSSE+LQIPE MV+LVIQRYTREAGVRN
Sbjct: 361 RMEIIELPGYTPEEKLHIAMRHLIPRVLDQHGLSSEFLQIPEAMVKLVIQRYTREAGVRN 540
Query: 598 LERNLASLARAAAVRVAEQEQVVPLNKEVQGLTTPLLENRLTDGAEVEMEVIPMDVNSRD 657
LERNLA+LARAAAV V EQEQVVPLNK +QGL TPL+ENRL DG EVEMEVIPM VNSRD
Sbjct: 541 LERNLAALARAAAVIVLEQEQVVPLNKGMQGLATPLVENRLADGTEVEMEVIPMGVNSRD 720
Query: 658 ISNTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEAT 717
IS+TFRI SPLVV+E +LEKVLGPP+FDGREAAERVA PG+ VGLVWT FGGEVQFVEAT
Sbjct: 721 ISSTFRIASPLVVDETMLEKVLGPPRFDGREAAERVATPGVSVGLVWTAFGGEVQFVEAT 900
Query: 718 EMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGA 777
MVGKG+LHLTGQLGDVIKESAQIALTWVRARA DLRLA+ +G N+LEGRD+HIHFPA A
Sbjct: 901 AMVGKGELHLTGQLGDVIKESAQIALTWVRARATDLRLAATKGFNILEGRDVHIHFPARA 1080
Query: 778 IPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIK 837
+PKDGPS GVTLVTALVSLFS++RVRSDTAMTGEMTL GLVLPVG +KDKIL H IK
Sbjct: 1081VPKDGPSTGVTLVTALVSLFSQQRVRSDTAMTGEMTLXGLVLPVGSVKDKILXTHHYNIK 1260
Query: 838 RVIIPERNLK 847
R+I+PE+NLK
Sbjct: 1261RIILPEKNLK 1290
>TC205454 similar to UP|LON1_ARATH (O64948) Lon protease homolog 1,
mitochondrial precursor , partial (25%)
Length = 1092
Score = 413 bits (1062), Expect = e-115
Identities = 207/229 (90%), Positives = 219/229 (95%)
Frame = +1
Query: 660 NTFRITSPLVVNEAILEKVLGPPKFDGREAAERVAAPGICVGLVWTTFGGEVQFVEATEM 719
NTFRITSPLVV+EA+LEKVLGPPKFDGREA +RVA PG VGLVWTTFGGEVQFVEAT M
Sbjct: 1 NTFRITSPLVVDEAMLEKVLGPPKFDGREAEDRVATPGASVGLVWTTFGGEVQFVEATAM 180
Query: 720 VGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIHIHFPAGAIP 779
VGKG+LHLTGQLGDVIKESAQIALTWVRARA +LRLA+AEGINLLEGRDIHIHFPAGA+P
Sbjct: 181 VGKGELHLTGQLGDVIKESAQIALTWVRARATELRLAAAEGINLLEGRDIHIHFPAGAVP 360
Query: 780 KDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRV 839
KDGPSAGVTLVTALVSLFS++RVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRV
Sbjct: 361 KDGPSAGVTLVTALVSLFSQRRVRSDTAMTGEMTLRGLVLPVGGIKDKILAAHRCGIKRV 540
Query: 840 IIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
I+PERNLKDLVEVPSSVL +LEILLAKRMEDVLEQA +GGCPWR SKL
Sbjct: 541 ILPERNLKDLVEVPSSVLADLEILLAKRMEDVLEQAFDGGCPWRQHSKL 687
>TC205452 similar to UP|LON1_ARATH (O64948) Lon protease homolog 1,
mitochondrial precursor , partial (19%)
Length = 682
Score = 310 bits (793), Expect = 2e-84
Identities = 155/178 (87%), Positives = 166/178 (93%)
Frame = +1
Query: 711 VQFVEATEMVGKGDLHLTGQLGDVIKESAQIALTWVRARAADLRLASAEGINLLEGRDIH 770
VQFVEAT MV KG+LHLTGQLGDVIKESAQIALTWVRAR DLRLA+ EG N+LEGRD+H
Sbjct: 1 VQFVEATAMVRKGELHLTGQLGDVIKESAQIALTWVRARTTDLRLAATEGFNILEGRDVH 180
Query: 771 IHFPAGAIPKDGPSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDKILA 830
IHFPAGA+PKDGPSAGVTLVTALVSLFS++RVRSDTAMTGEMTLRGLVLPVGG+KDKILA
Sbjct: 181 IHFPAGAVPKDGPSAGVTLVTALVSLFSQQRVRSDTAMTGEMTLRGLVLPVGGVKDKILA 360
Query: 831 AHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRHQSKL 888
AHR GIKRVI+PERNLKDLVEVPSSVL NLEILLAKR+EDVLE A +GGCPWR SKL
Sbjct: 361 AHRYGIKRVILPERNLKDLVEVPSSVLSNLEILLAKRVEDVLEHAFDGGCPWRQHSKL 534
>TC205450 similar to UP|Q8GV57 (Q8GV57) Lon protease, partial (12%)
Length = 1003
Score = 150 bits (380), Expect = 2e-36
Identities = 88/139 (63%), Positives = 96/139 (68%), Gaps = 33/139 (23%)
Frame = +2
Query: 783 PSAGVTLVTALVSLFSKKRVRSDTAMTGEMTLRGLVLPVGGIKDK--------------- 827
PSA V L ++VSLFS++RVRSDTAMTGEMTLRGLVLPVGG+KDK
Sbjct: 23 PSA-V*LGHSIVSLFSQQRVRSDTAMTGEMTLRGLVLPVGGVKDKVKFYIFLTFVNFLRQ 199
Query: 828 ------------------ILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRME 869
ILAAHR GIKRVI+PERNLKDLVEVPSSVL NLEILLAKR+E
Sbjct: 200 DK*LLIE*IQSLGGVKYKILAAHRYGIKRVILPERNLKDLVEVPSSVLSNLEILLAKRVE 379
Query: 870 DVLEQALEGGCPWRHQSKL 888
DVLE A +GGCPWR SKL
Sbjct: 380 DVLEHAFDGGCPWRQHSKL 436
>BQ253435 similar to SP|O04979|LON1_ Lon protease homolog 1 mitochondrial
precursor (EC 3.4.21.-). [Spinach] {Spinacia oleracea},
partial (6%)
Length = 421
Score = 150 bits (380), Expect = 2e-36
Identities = 80/101 (79%), Positives = 87/101 (85%), Gaps = 2/101 (1%)
Frame = +2
Query: 1 MAESVELPSRLAILPFRNKVLLPGAIIRIRCTSPTSVKLVEQELWQREDKGLIGILPVRD 60
MAES ELP+RLAILPFRNKVLLPGAIIRIRCTSP SVKLVEQELWQRE+KGLIGILPVRD
Sbjct: 119 MAESFELPNRLAILPFRNKVLLPGAIIRIRCTSPISVKLVEQELWQREEKGLIGILPVRD 298
Query: 61 AAETEVKPVGPTIS--QDGDTLDQSSKVHGASSDSHKLDAK 99
AA E++P GP IS + D+LDQ+SKV G SSDS KLD K
Sbjct: 299 AA-AEIQPAGPVISHGKGTDSLDQNSKVQGGSSDSQKLDVK 418
>BI972574 homologue to SP|P93655|LON2_ Lon protease homolog 2 mitochondrial
precursor (EC 3.4.21.-). [Mouse-ear cress], partial
(10%)
Length = 271
Score = 121 bits (304), Expect = 1e-27
Identities = 57/90 (63%), Positives = 70/90 (77%)
Frame = +2
Query: 443 DIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLDPEQN 502
+I+GHRRTYIG+MPG+++ LK V NP++L+DEIDK G GDPASALLE+LDPEQN
Sbjct: 2 EIKGHRRTYIGAMPGKIVQCLKNVGTSNPLVLIDEIDKLGRGHAGDPASALLELLDPEQN 181
Query: 503 KTFNDHYLNVPFDLSKVIFVATANRAQQIP 532
F DHYL+V DLSKV+FV TAN + IP
Sbjct: 182 ANFLDHYLDVTIDLSKVLFVCTANVVEMIP 271
>TC205451 similar to UP|Q8GV57 (Q8GV57) Lon protease, partial (7%)
Length = 641
Score = 111 bits (277), Expect = 2e-24
Identities = 54/64 (84%), Positives = 58/64 (90%)
Frame = +1
Query: 825 KDKILAAHRCGIKRVIIPERNLKDLVEVPSSVLVNLEILLAKRMEDVLEQALEGGCPWRH 884
KDKILAAHR GIKRVI+PERNLKDLVEVPSSVL NLE+LLAKR+EDVLE A +GGCPWR
Sbjct: 1 KDKILAAHRYGIKRVILPERNLKDLVEVPSSVLSNLEVLLAKRVEDVLEHAFDGGCPWRQ 180
Query: 885 QSKL 888
SKL
Sbjct: 181 HSKL 192
>AI437651 similar to SP|O04979|LON1_ Lon protease homolog 1 mitochondrial
precursor (EC 3.4.21.-). [Spinach] {Spinacia oleracea},
partial (5%)
Length = 140
Score = 81.6 bits (200), Expect = 1e-15
Identities = 41/46 (89%), Positives = 43/46 (93%)
Frame = +2
Query: 194 VLEQKQKTGGRTKVLLETVPVHKLADIFVASFEISFEEQLSMLDSV 239
VLE KQKTGGRTKVLL+ VPVHKLADIFVASFEISFEEQLSMLD +
Sbjct: 2 VLELKQKTGGRTKVLLDNVPVHKLADIFVASFEISFEEQLSMLDLI 139
>BM308386 similar to SP|P93648|LON2 Lon protease homolog 2 mitochondrial
precursor (EC 3.4.21.-). [Maize] {Zea mays}, partial
(11%)
Length = 335
Score = 68.6 bits (166), Expect = 1e-11
Identities = 35/91 (38%), Positives = 56/91 (61%)
Frame = +2
Query: 316 PQSIWKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPWQKASEEHEMDLKSAQERLDS 375
P I + EL +L+ ++ +S +R YLD L LPW + S+E+ D+ + LD
Sbjct: 59 PPHILQVIDEELAKLQLLEASSSEFSVTRNYLDWLTALPWGEYSDEN-FDVTRL*KILDE 235
Query: 376 DHYGLAKVKQRIIEYLAVRKLKPDARGPVLC 406
DHYGL VK+RI+E++AV KL+ ++G ++C
Sbjct: 236 DHYGLTDVKERILEFIAVGKLRGTSQGKIIC 328
>BF070462 similar to SP|P93648|LON2 Lon protease homolog 2 mitochondrial
precursor (EC 3.4.21.-). [Maize] {Zea mays}, partial
(10%)
Length = 418
Score = 53.5 bits (127), Expect = 4e-07
Identities = 31/96 (32%), Positives = 50/96 (51%), Gaps = 3/96 (3%)
Frame = +1
Query: 263 EKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDDDEDDLAALERKM---QSAGMPQSI 319
E I + +E ++S Q+ +LL + + AIK+ELG DD+ L R+ + P I
Sbjct: 10 ESIAKSIEEKISGEQRSYLLNEHLNAIKKELGLETDDKTALTGKFRERIEPKREKCPPHI 189
Query: 320 WKHAHRELRRLKKMQPQQPGYSSSRAYLDLLADLPW 355
+ EL +L+ ++ +S +R YLD L LPW
Sbjct: 190 LQVIDEELAKLQLLEASSSEFSVTRNYLDWLTALPW 297
>TC215774 similar to UP|Q94ES0 (Q94ES0) AAA-metalloprotease FtsH, partial
(79%)
Length = 2084
Score = 45.4 bits (106), Expect = 1e-04
Identities = 53/186 (28%), Positives = 79/186 (41%), Gaps = 16/186 (8%)
Frame = +3
Query: 376 DHYGLAKVKQRIIEYLAVRK--LKPDARGPVL----CFVGPPGVGKTSLASSIAAALDRK 429
D G + KQ I+E++ K K + G + VGPPG GKT LA + A
Sbjct: 501 DVAGCDEAKQEIMEFVHFLKNPKKYEELGAKIPKGALLVGPPGTGKTLLAKATAGESGVP 680
Query: 430 FVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VMLLDEIDKTGSDVRGD 488
F+ IS + ++G P R+ + + C+P ++ +DEID G RG
Sbjct: 681 FLSISGSDFME---------MFVGVGPSRVRNLFQEARQCSPSIVFIDEIDAIGRARRGS 833
Query: 489 PASALLEVLDPEQNKTFNDHYLNVPFD----LSKVIFVATANRAQQIPPPLL-----DRM 539
+ A + E+ T N L V D S V+ +A NR + + LL DR
Sbjct: 834 FSGA-----NDERESTLNQ--LLVEMDGFGTTSGVVVLAGTNRPEILDKALLRPGRFDRQ 992
Query: 540 EVIELP 545
I+ P
Sbjct: 993 ITIDKP 1010
>TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulatory particle
triple-A ATPase subunit4, partial (62%)
Length = 1079
Score = 43.1 bits (100), Expect = 5e-04
Identities = 61/220 (27%), Positives = 102/220 (45%), Gaps = 8/220 (3%)
Frame = +3
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA +IA+ ++ F+++ + D+ YIG RLI + A
Sbjct: 99 GPPGTGKTLLARAIASNIEANFLKVVSSAIIDK---------YIGE-SARLIREMFGYAR 248
Query: 469 CNP--VMLLDEIDKTGSDVRGDPASALLEVLDPEQNKTFND--HYLNVPFDLSKVIFVAT 524
+ ++ +DEID G + SA D E +T + + L+ L KV +
Sbjct: 249 DHQPCIIFMDEIDAIGGRRFSEGTSA-----DREIQRTLMELLNQLDGFDQLGKVKMIMA 413
Query: 525 ANRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEGM 581
NR + P LL R++ IE+P P E+ ++ + + + +HG +Y E +
Sbjct: 414 TNRPDVLDPALLRPGRLDRKIEIP--LPNEQSRMEILKIHAAGIAKHG-EIDY----EAV 572
Query: 582 VQLVIQRYTREAGVRNLE-RNLASLARAAAVRVAEQEQVV 620
V+L G + RN+ + A AA+R AE++ V+
Sbjct: 573 VKLA-------EGFNGADLRNVCTEAGMAAIR-AERDYVI 668
>TC214213 homologue to UP|PRSA_BRACM (O23894) 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1),
partial (64%)
Length = 1086
Score = 40.8 bits (94), Expect = 0.003
Identities = 56/217 (25%), Positives = 94/217 (42%), Gaps = 20/217 (9%)
Frame = +1
Query: 363 EMDLKSAQERLDSDHYGLAKVKQRIIEYLAV------RKLKPDARGPV-LCFVGPPGVGK 415
E+D K ++ +D GL K Q ++E + + R K R P + GPPG GK
Sbjct: 19 EVDEKPTEDY--NDIGGLEKQIQELVEAIVLPMTHKERFQKLGVRPPKGVLLYGPPGTGK 192
Query: 416 TSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VML 474
T +A + AA + F++++ + + +IG + D + +P ++
Sbjct: 193 TLMARACAAQTNATFLKLAGPQLV---------QMFIGDGAKLVRDAFQLAKEKSPCIIF 345
Query: 475 LDEIDKTG-----SDVRGD--PASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANR 527
+DEID G S+V GD +LE+L+ + +D ++ +A NR
Sbjct: 346 IDEIDAIGTKRFDSEVSGDREVQRTMLELLNQLDGFSSDD----------RIKVIAATNR 495
Query: 528 AQQIPPPL-----LDRMEVIELPGYTPEEKLKIAMKH 559
A + P L LDR IE P + E + +I H
Sbjct: 496 ADILDPALMRSGRLDRK--IEFPHPSEEARARILQIH 600
>NP005140 heat shock protein
Length = 2736
Score = 40.0 bits (92), Expect = 0.005
Identities = 32/99 (32%), Positives = 52/99 (52%), Gaps = 7/99 (7%)
Frame = +1
Query: 407 FVGPPGVGKTSLASSIAAAL---DRKFVRISLGGVKDEADIR---GHRRTYIG-SMPGRL 459
F+GP GVGKT LA ++A L + + VRI + ++ + G Y+G G+L
Sbjct: 1810 FLGPTGVGKTELAKALAEQLFDNENQLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEGGQL 1989
Query: 460 IDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLD 498
+ ++R V+L DE++K + V + LL+VLD
Sbjct: 1990 TEAVRRRPY--SVVLFDEVEKAHTSV----FNTLLQVLD 2088
>TC206564 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (40%)
Length = 1497
Score = 39.3 bits (90), Expect = 0.008
Identities = 33/99 (33%), Positives = 53/99 (53%), Gaps = 7/99 (7%)
Frame = +2
Query: 407 FVGPPGVGKTSLASSIAAAL---DRKFVRISLGGVKDE---ADIRGHRRTYIG-SMPGRL 459
F+GP GVGKT LA ++AA L + VRI + ++ + + G Y+G G+L
Sbjct: 374 FMGPTGVGKTELAKALAAYLFNTEEALVRIDMSEYMEKHAVSRLIGAPPGYVGYEEGGQL 553
Query: 460 IDGLKRVAVCNPVMLLDEIDKTGSDVRGDPASALLEVLD 498
+ ++R V+L DEI+K +DV + L++LD
Sbjct: 554 TEIVRRRPYA--VILFDEIEKAHADV----FNVFLQILD 652
>TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (38%)
Length = 1502
Score = 38.9 bits (89), Expect = 0.010
Identities = 48/220 (21%), Positives = 94/220 (41%), Gaps = 14/220 (6%)
Frame = +2
Query: 375 SDHYGLAKVKQRIIEYLAVRKLKPDA-RGPVL------CFVGPPGVGKTSLASSIAAALD 427
+D L ++K+ + E + + +PD +G +L GPPG GKT LA +IA
Sbjct: 62 ADIGALDEIKESLQELVMLPLRRPDLFKGGLLKPCRGILLFGPPGTGKTMLAKAIANEAG 241
Query: 428 RKFVRISLGGV------KDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNPVMLLDEIDK- 480
F+ +S+ + +DE ++R + ++ + V ++ +DE+D
Sbjct: 242 ASFINVSMSTITSKWFGEDEKNVRA-----LFTLAAK---------VAPTIIFVDEVDSM 379
Query: 481 TGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLLDRME 540
G R A+ ++ + T D L P + +++ +A NR + ++ R E
Sbjct: 380 LGQRTRVGEHEAMRKI--KNEFMTHWDGLLTGPNE--QILVLAATNRPFDLDEAIIRRFE 547
Query: 541 VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEYLQIPEG 580
L G E ++ +K L+ + ++ E + EG
Sbjct: 548 RRILVGLPSVENREMILKTLLAKEKHENLDFKELATMTEG 667
>TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 homolog
(Valosin containing protein homolog) (VCP), complete
Length = 2583
Score = 38.5 bits (88), Expect = 0.013
Identities = 57/214 (26%), Positives = 88/214 (40%), Gaps = 18/214 (8%)
Frame = +1
Query: 407 FVGPPGVGKTSLASSIAAALDRKFVRI------SLGGVKDEADIRGHRRTYIGSMPGRLI 460
F GPPG GKT LA +IA F+ + ++ + EA++R +
Sbjct: 1579 FYGPPGCGKTLLAKAIANECQANFISVKGPELLTMWFGESEANVR------------EIF 1722
Query: 461 DGLKRVAVCNPVMLLDEID----KTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDL 516
D ++ A C V+ DE+D + GS V GD A VL+ Q T D
Sbjct: 1723 DKARQSAPC--VLFFDELDSIATQRGSSV-GDAGGAADRVLN--QLLTEMD-----GMSA 1872
Query: 517 SKVIFVATA-NRAQQIPPPL-----LDRMEVIELPGYTPEEKLKIA--MKHLIPRVLDQH 568
K +F+ A NR I P L LD++ I LP ++ A K I + +D
Sbjct: 1873 KKTVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEDSRHQIFKACLRKSPIAKNVDLR 2052
Query: 569 GLSSEYLQIPEGMVQLVIQRYTREAGVRNLERNL 602
L+ + + QR + A N+E+++
Sbjct: 2053 ALARHTQGFSGADITEICQRACKYAIRENIEKDI 2154
>TC205434 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regulatory subunit
6B homolog (26S proteasome AAA-ATPase subunit RPT3)
(Regulatory particle triple-A ATPase subunit 3), partial
(96%)
Length = 1573
Score = 37.7 bits (86), Expect = 0.022
Identities = 57/207 (27%), Positives = 91/207 (43%), Gaps = 14/207 (6%)
Frame = +1
Query: 409 GPPGVGKTSLASSIAAALDRKFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAV 468
GPPG GKT LA ++A F+R+ V E + Y+G P R++ + R+A
Sbjct: 700 GPPGTGKTMLAKAVANHTTAAFIRV----VGSE-----FVQKYLGEGP-RMVRDVFRLAK 849
Query: 469 CN--PVMLLDEID---------KTGSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLS 517
N ++ +DE+D +TG+D + L+E+L+ Q F D +NV
Sbjct: 850 ENAPAIIFIDEVDAIATARFDAQTGAD--REVQRILMELLN--QMDGF-DQTVNV----- 999
Query: 518 KVIFVATANRAQQIPPPLL--DRME-VIELPGYTPEEKLKIAMKHLIPRVLDQHGLSSEY 574
KVI NRA + P LL R++ IE P +K + L +Y
Sbjct: 1000KVIM--ATNRADTLDPALLRPGRLDRKIEFPLPDRRQKRLVFQVCTAKMNLSDEVDLEDY 1173
Query: 575 LQIPEGMVQLVIQRYTREAGVRNLERN 601
+ P+ + I +EAG+ + +N
Sbjct: 1174VSRPDKISAAEIAAICQEAGMHAVRKN 1254
>TC205253 homologue to UP|O99018 (O99018) Chloroplast protease precursor,
partial (69%)
Length = 1678
Score = 37.0 bits (84), Expect = 0.038
Identities = 50/193 (25%), Positives = 76/193 (38%), Gaps = 23/193 (11%)
Frame = +2
Query: 376 DHYGLAKVKQRIIEYLAVRKLKPD------ARGPV-LCFVGPPGVGKTSLASSIAAALDR 428
D G+ + KQ +E + K KP+ AR P + VGPPG GKT LA +IA
Sbjct: 62 DVAGVDEAKQDFMEVVEFLK-KPERFTAVGARIPKGVLLVGPPGTGKTLLAKAIAGEAGV 238
Query: 429 KFVRISLGGVKDEADIRGHRRTYIGSMPGRLIDGLKRVAVCNP-VMLLDEIDKT------ 481
F IS + ++G R+ D ++ P ++ +DEID
Sbjct: 239 PFFSISGSEFVE---------MFVGVGASRVRDLFRKAKENAPCIVFVDEIDAVGRQRGT 391
Query: 482 ----GSDVRGDPASALLEVLDPEQNKTFNDHYLNVPFDLSKVIFVATANRAQQIPPPLL- 536
G+D R + LL +D + T +I +A NR + LL
Sbjct: 392 GIGGGNDEREQTLNQLLTEMDGFEGNT-------------GIIVIAATNRVDILDSALLR 532
Query: 537 ----DRMEVIELP 545
DR +++P
Sbjct: 533 PGRFDRQVTVDVP 571
>TC206435 homologue to UP|PRSA_LYCES (P54776) 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1)
(Mg(2+)-dependent ATPase 1) (LEMA-1), partial (61%)
Length = 838
Score = 36.6 bits (83), Expect = 0.050
Identities = 51/211 (24%), Positives = 92/211 (43%), Gaps = 15/211 (7%)
Frame = +2
Query: 239 VDAKLRLSKATELVDRHLQSIRVAEKITQKVEGQLSKSQKEFLLRQQMRAIKEELGDNDD 298
+D ++R+ K EL +L+ +KI + E Q +L+ + + E+ D+
Sbjct: 152 LDNEIRILKE-ELQRTNLELESYKDKIKENQEKIKLNKQLPYLVGNIVEIL--EMNPEDE 322
Query: 299 DEDDLAALERKMQSAGMPQSIWKHAHRELRRLK--------KMQPQQPGYSSSRAYLDLL 350
E+D A ++ Q G + K + R+ L K++P + +YL +L
Sbjct: 323 AEEDGANIDLDSQRKGKCV-VLKTSTRQTIFLPVVGLVDPDKLKPGDLVGVNKDSYL-IL 496
Query: 351 ADLPWQKASEEHEMDLKSAQERLDSDHYGLAKVKQRIIEYLAV------RKLKPDARGPV 404
LP + S M++ +D GL K Q ++E + + R K R P
Sbjct: 497 DTLPSEYDSRVKAMEVDEKPTEDYNDIGGLEKQIQELVEAIVLPMTCKERFQKLGVRPPK 676
Query: 405 -LCFVGPPGVGKTSLASSIAAALDRKFVRIS 434
+ GPPG GKT +A + AA + F++++
Sbjct: 677 GVLLYGPPGTGKTLIARACAAQTNATFLKLA 769
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,098,512
Number of Sequences: 63676
Number of extensions: 364861
Number of successful extensions: 1833
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 1778
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1809
length of query: 888
length of database: 12,639,632
effective HSP length: 106
effective length of query: 782
effective length of database: 5,889,976
effective search space: 4605961232
effective search space used: 4605961232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0550.4


