

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
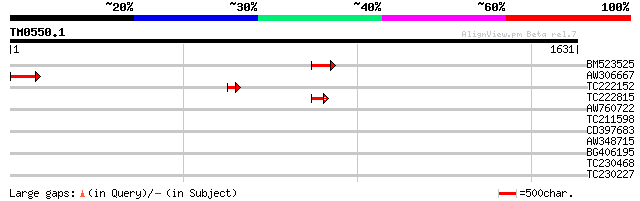
Query= TM0550.1
(1631 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BM523525 128 2e-29
AW306667 homologue to PIR|B96521|B965 protein F21D18.20 [importe... 123 8e-28
TC222152 59 2e-08
TC222815 similar to UP|NDI1_YEAST (P32340) Rotenone-insensitive ... 52 2e-06
AW760722 36 0.16
TC211598 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At... 33 1.3
CD397683 32 3.0
AW348715 30 6.6
BG406195 30 6.6
TC230468 similar to UP|Q9FHS2 (Q9FHS2) Gb|AAF03435.1, partial (11%) 30 8.7
TC230227 30 8.7
>BM523525
Length = 429
Score = 128 bits (322), Expect = 2e-29
Identities = 61/70 (87%), Positives = 67/70 (95%)
Frame = +2
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVEFAASLHD+V EDLV +YPGIKD+VKITLLEAGDHILSMFDKRITAFA++KF R
Sbjct: 218 GGPTGVEFAASLHDYVTEDLVNIYPGIKDLVKITLLEAGDHILSMFDKRITAFAEEKFGR 397
Query: 927 DGIDVKTGSV 936
DGIDVKTGS+
Sbjct: 398 DGIDVKTGSM 427
>AW306667 homologue to PIR|B96521|B965 protein F21D18.20 [imported] -
Arabidopsis thaliana, partial (18%)
Length = 428
Score = 123 bits (308), Expect = 8e-28
Identities = 58/89 (65%), Positives = 74/89 (82%)
Frame = +2
Query: 1 MLEDQVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGF 60
M E V +LL+ YLG YV GL+ EAL+ISVWKGDV LK+++LK EALNALKLPV VKAGF
Sbjct: 161 MFEAHVLHLLRSYLGEYVHGLSTEALRISVWKGDVVLKDLKLKAEALNALKLPVTVKAGF 340
Query: 61 LGSVKLKVPWSRLGQDPVLVYLDRIFLLA 89
+G++ LKVPW LG++PV+V +DR+F+LA
Sbjct: 341 VGTITLKVPWKSLGKEPVIVLIDRVFVLA 427
>TC222152
Length = 1018
Score = 58.5 bits (140), Expect = 2e-08
Identities = 29/36 (80%), Positives = 32/36 (88%)
Frame = +1
Query: 627 YMKDSINQIVKFFETNATVSQTIALETAAAVQLKID 662
YMKDSI+QIVKFFE+N VSQTIALETAAAVQ+ D
Sbjct: 31 YMKDSIDQIVKFFESNTAVSQTIALETAAAVQVLFD 138
>TC222815 similar to UP|NDI1_YEAST (P32340) Rotenone-insensitive
NADH-ubiquinone oxidoreductase, mitochondrial precursor
(Internal NADH dehydrogenase) , partial (8%)
Length = 612
Score = 52.4 bits (124), Expect = 2e-06
Identities = 25/50 (50%), Positives = 35/50 (70%)
Frame = +3
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRI 916
GGPTGVEF+ L DF+ D+ + Y +KD + +TL+EA + ILS FD R+
Sbjct: 465 GGPTGVEFSGELSDFIVRDVRQRYAHVKDYIHVTLIEANE-ILSSFDVRL 611
>AW760722
Length = 391
Score = 35.8 bits (81), Expect = 0.16
Identities = 17/45 (37%), Positives = 27/45 (59%)
Frame = +2
Query: 1137 IGLLPTDAPSTSTKEHELNDTLESFVKAQIIIYDQNSTRYNNIDK 1181
IGL P P + ++ +N E VK +I +Y+QN ++ NN+DK
Sbjct: 62 IGLYPDILPDLAEEDDAIN---EDIVKLEIALYEQNGSQKNNLDK 187
>TC211598 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;} , partial (13%)
Length = 599
Score = 32.7 bits (73), Expect = 1.3
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Frame = +3
Query: 1168 IYDQNSTRYNNIDKQVIVTLATLTFFC------RRPTILAIMEFINSINIENGNLATSSD 1221
++DQ RY NI+++ +V++ + C +RPT+ +++ I I +E L D
Sbjct: 207 VFDQELLRYKNIEEE-LVSMLHVGLTCVVAQPEKRPTMEEVVKMIEEIRVEQSPLGEDYD 383
Query: 1222 SSSTSM 1227
S S+
Sbjct: 384 VSCNSL 401
>CD397683
Length = 591
Score = 31.6 bits (70), Expect = 3.0
Identities = 11/34 (32%), Positives = 24/34 (70%)
Frame = +3
Query: 794 YPSTRLAVRLPSLAFHFSPARYHRLMHVIKIFEE 827
+PST+ +L +F +P+R H+++H++K ++E
Sbjct: 27 FPSTKSTQKLNGTSFFSNPSR*HQILHLVKPYKE 128
>AW348715
Length = 713
Score = 30.4 bits (67), Expect = 6.6
Identities = 23/100 (23%), Positives = 47/100 (47%), Gaps = 12/100 (12%)
Frame = -1
Query: 1136 IIGLLPTDAPSTSTKEHELNDTLESFVKAQII------IYDQNSTRYNNIDKQVIVTLAT 1189
+ G P+ ++ +E E++ L +VK+ + ++DQ RY NI+ + +V +
Sbjct: 467 LTGRAPSKEYTSPAREAEVD--LPKWVKSVVKEEWTSEVFDQELLRYKNIEDE-LVAMLH 297
Query: 1190 LTFFC------RRPTILAIMEFINSINIENGNLATSSDSS 1223
+ C +RP +L +++ I I +E L D +
Sbjct: 296 VGLACVAAQAEKRPCMLEVVKMIEEIRVEESPLGDDYDEA 177
>BG406195
Length = 328
Score = 30.4 bits (67), Expect = 6.6
Identities = 13/28 (46%), Positives = 18/28 (63%)
Frame = +2
Query: 1588 VEASVLLKIWVSINLVELSLYTGISRDA 1615
V VLLKIW S+ L+ ++L G + DA
Sbjct: 110 VTPMVLLKIWYSVTLININLTNGFTMDA 193
>TC230468 similar to UP|Q9FHS2 (Q9FHS2) Gb|AAF03435.1, partial (11%)
Length = 707
Score = 30.0 bits (66), Expect = 8.7
Identities = 25/95 (26%), Positives = 39/95 (40%)
Frame = -1
Query: 1057 SFIGAADEKSLFYDTMRENVESSGLIPTESDDKFYEAPETLADSDVYMQSPGGTSEYPSS 1116
S + A KS + T+R + S+ ESD+ E+ E SS
Sbjct: 557 SLVFHASRKSSYRTTLRPPLTSASFSLDESDESSIPRAES--------------GESNSS 420
Query: 1117 SSNEIKFNYSSLEPPKFSRIIGLLPTDAPSTSTKE 1151
SS+ + +N+ S P S + L PT +T+E
Sbjct: 419 SSSSLVYNFLSTNDPVLSEV--LFPTSCSLPTTEE 321
>TC230227
Length = 781
Score = 30.0 bits (66), Expect = 8.7
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Frame = -1
Query: 493 SEELSFSEED-WNRLNKIIGYKEGDDGQSPVNSKADVMHTFLVV 535
SE+LSFSEED W ++ ++ G+ SP + HTF ++
Sbjct: 379 SEKLSFSEEDPWPQIKEVCGFVLPSSSPSP*PHTST*HHTFKIL 248
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,524,370
Number of Sequences: 63676
Number of extensions: 847671
Number of successful extensions: 4130
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 4078
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4128
length of query: 1631
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1521
effective length of database: 5,635,272
effective search space: 8571248712
effective search space used: 8571248712
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0550.1


