

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0546.10
(175 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
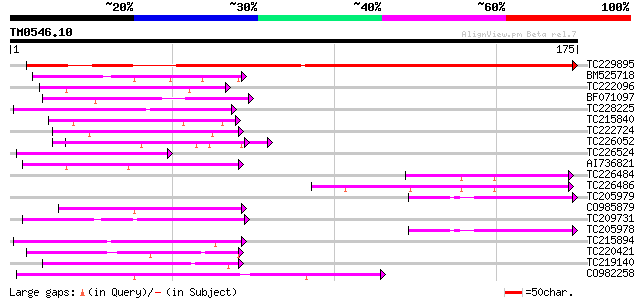
Score E
Sequences producing significant alignments: (bits) Value
TC229895 homologue to UP|Q7YAY7 (Q7YAY7) Cytochrome b (Fragment)... 219 5e-58
BM525718 similar to PIR|T04592|T045 glycine-rich cell wall struc... 45 1e-05
TC222096 44 3e-05
BF071097 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 44 5e-05
TC228225 weakly similar to UP|Q9FFJ7 (Q9FFJ7) Similarity to C3HC... 44 5e-05
TC215840 homologue to UP|Q41122 (Q41122) Proline-rich protein pr... 43 9e-05
TC222724 weakly similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (5%) 42 1e-04
TC226052 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_... 42 1e-04
TC226524 similar to UP|Q8VYJ5 (Q8VYJ5) At1g68530/T26J14_10, part... 42 1e-04
AI736821 weakly similar to GP|11036868|gb| PxORF73 peptide {Plut... 42 1e-04
TC226484 similar to GB|AAP21355.1|30102874|BT006547 At1g75690 {A... 42 2e-04
TC226486 similar to GB|AAP21355.1|30102874|BT006547 At1g75690 {A... 41 3e-04
TC205979 homologue to UP|Q6XPS8 (Q6XPS8) Drought-induced protein... 41 3e-04
CO985879 41 3e-04
TC209731 similar to UP|Q8W3M8 (Q8W3M8) 6b-interacting protein 1,... 41 3e-04
TC205978 homologue to UP|Q6XPS8 (Q6XPS8) Drought-induced protein... 41 3e-04
TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase ... 41 3e-04
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 41 3e-04
TC219140 similar to GB|AAO44047.1|28466877|BT004781 At2g39020 {A... 41 3e-04
CO982258 40 4e-04
>TC229895 homologue to UP|Q7YAY7 (Q7YAY7) Cytochrome b (Fragment), partial
(5%)
Length = 666
Score = 219 bits (558), Expect = 5e-58
Identities = 117/170 (68%), Positives = 122/170 (70%)
Frame = +2
Query: 6 YSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCR 65
Y TLHPLSSS P SS S+HL PCSS + SLP+FPSK R
Sbjct: 17 YYTLHPLSSSLTP-------SSKGSVHLCPCSS-------------TRSRSLPQFPSKLR 136
Query: 66 HKPPKPPRLFMIIDPILLLNGFGGAGFYLDTQTLLATVSVLAAIALSLFLGLKGDPVPCE 125
K PRL IIDPILL NG FY DTQTL+ TVSVLAAIALSLFLGLKGDPVPCE
Sbjct: 137 RKATIKPRLLTIIDPILLFNGVATT-FYFDTQTLIVTVSVLAAIALSLFLGLKGDPVPCE 313
Query: 126 RCAGNGGMKCVFCNDGKMKQEMGLIDCKVCKGSGLILCKKCGGSGYSRRL 175
RC GNGG KCVFCNDGKMKQEMGLI+CKVCKGSGLI CKKCGGSGYSRRL
Sbjct: 314 RCGGNGGTKCVFCNDGKMKQEMGLINCKVCKGSGLIFCKKCGGSGYSRRL 463
>BM525718 similar to PIR|T04592|T045 glycine-rich cell wall structural
protein homolog F23E13.120 - Arabidopsis thaliana,
partial (11%)
Length = 326
Score = 45.4 bits (106), Expect = 1e-05
Identities = 35/74 (47%), Positives = 38/74 (51%), Gaps = 8/74 (10%)
Frame = +2
Query: 8 TLHPLSSSSLPPSSSSSSSSSTSLHLSPCS-SHSLPQPSPPP-PSKSHFPSLP--RFPSK 63
T H S + PSSSSSSSSS+ H P S H L P PPP P SH P P R P
Sbjct: 89 TQHHKYSMPMVPSSSSSSSSSS--HTPPSS*PHHLLHPLPPPHPGPSHLPLRPPSRAPGL 262
Query: 64 CRHKPP----KPPR 73
RH+ P PPR
Sbjct: 263 RRHQSPPNRSPPPR 304
Score = 26.6 bits (57), Expect = 6.4
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 4/47 (8%)
Frame = +1
Query: 12 LSSSSLPPSSSSSSSSSTSLHLSPCSSHSLP----QPSPPPPSKSHF 54
L +S PP + SSS S S P S P +PS P PS + F
Sbjct: 169 LLTSPPPPPAPSSSPRSLSPPTPPSEPSSRPSETSEPSKPKPSATAF 309
>TC222096
Length = 728
Score = 44.3 bits (103), Expect = 3e-05
Identities = 29/65 (44%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Frame = +2
Query: 10 HPLSSSS-LPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHF-----PSLPRFPSK 63
HPLS SS PPS +SS SST +P S S P PPP S + P PR PS
Sbjct: 203 HPLSFSSYFPPSPLASSPSSTETTATPLSPPSPTTPPPPPAS*TLLIPPPPPPSPRSPSS 382
Query: 64 CRHKP 68
+H P
Sbjct: 383 TQHHP 397
Score = 26.2 bits (56), Expect = 8.4
Identities = 18/42 (42%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Frame = +1
Query: 3 TLSYSTLHPLSSSS-LPPSSSSSSSSSTSLHLSPCSSHSLPQ 43
+LS T P ++S L PS+SSSSSS ++L L +S P+
Sbjct: 283 SLSSFTYDPTTTSCVLNPSNSSSSSSVSTLSLVDTTSSVSPK 408
>BF071097 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (18%)
Length = 366
Score = 43.5 bits (101), Expect = 5e-05
Identities = 31/69 (44%), Positives = 35/69 (49%), Gaps = 4/69 (5%)
Frame = +2
Query: 11 PLSSSSLPPSSSSSS----SSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRH 66
PLSS PPSS SS SSS L S C+S + P PSPP SLP PS
Sbjct: 134 PLSSKP*PPSSPPSSKPPASSSPPLPYSSCASTTHPPPSPP-------RSLPPPPSPTPP 292
Query: 67 KPPKPPRLF 75
P + PR+F
Sbjct: 293 SPRRKPRVF 319
Score = 32.7 bits (73), Expect = 0.090
Identities = 22/44 (50%), Positives = 22/44 (50%), Gaps = 4/44 (9%)
Frame = +2
Query: 34 SPCSSHSLPQPSPPPPSKSHFPSLPRFP-SKC---RHKPPKPPR 73
SP SS P PS PP SK S P P S C H PP PPR
Sbjct: 131 SPLSSKP*P-PSSPPSSKPPASSSPPLPYSSCASTTHPPPSPPR 259
Score = 28.9 bits (63), Expect = 1.3
Identities = 20/52 (38%), Positives = 22/52 (41%), Gaps = 7/52 (13%)
Frame = +3
Query: 29 TSLHLSPCSSHSLPQPSPPPPSKSHFP----SLPRFPSKC---RHKPPKPPR 73
T L P SH LP PSPP P+ P L R +C PP PR
Sbjct: 90 TLLPKPPLVSHPLPPPSPPNPNLLPLPRRRNHLHRHRRRCLILHALPPHTPR 245
Score = 26.2 bits (56), Expect = 8.4
Identities = 12/29 (41%), Positives = 14/29 (47%)
Frame = +2
Query: 44 PSPPPPSKSHFPSLPRFPSKCRHKPPKPP 72
P PPPP + PS P + R PP P
Sbjct: 50 PLPPPPRLGYTPSDPPPKTTPRFAPPPSP 136
>TC228225 weakly similar to UP|Q9FFJ7 (Q9FFJ7) Similarity to C3HC4-type
RING zinc finger protein, partial (10%)
Length = 689
Score = 43.5 bits (101), Expect = 5e-05
Identities = 28/69 (40%), Positives = 38/69 (54%)
Frame = +2
Query: 2 ATLSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP 61
++ S+S SSS+ PPSSSS+SSSS + + S+ LP P PPPS S P
Sbjct: 26 SSCSFSPPSSSSSSASPPSSSSTSSSSVAPSTAFASAPPLP-PRAPPPSASPLVISTTSP 202
Query: 62 SKCRHKPPK 70
+ C K P+
Sbjct: 203ALCSPKDPQ 229
>TC215840 homologue to UP|Q41122 (Q41122) Proline-rich protein precursor,
partial (74%)
Length = 1171
Score = 42.7 bits (99), Expect = 9e-05
Identities = 29/71 (40%), Positives = 34/71 (47%), Gaps = 12/71 (16%)
Frame = +1
Query: 13 SSSSLP-PSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKS---------HFPSLPRFPS 62
SS+ LP PS S +STS S CSS + P P PPP S S P LP +P
Sbjct: 142 SSTLLPSPSPCSLPHTSTSSPTSKCSSPTPPPPPPPPSSTSTSTSTCSFTFSPQLPSYPC 321
Query: 63 KC--RHKPPKP 71
C + PP P
Sbjct: 322 PC*ASYSPPSP 354
Score = 29.6 bits (65), Expect = 0.76
Identities = 23/69 (33%), Positives = 27/69 (38%), Gaps = 6/69 (8%)
Frame = +2
Query: 10 HPLSSSSLP---PSSSSSSSSSTSLHLSPCSSHSL---PQPSPPPPSKSHFPSLPRFPSK 63
HP + + P PS S + T P + H P P PPP H P P P
Sbjct: 248 HPPAPAPAPAPVPSPSHHNYPPTPAPAKPPTHHHHHHPPAPVNPPPVPVHPPVKPPVPV- 424
Query: 64 CRHKPPKPP 72
H P KPP
Sbjct: 425 --HPPVKPP 445
>TC222724 weakly similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (5%)
Length = 714
Score = 42.4 bits (98), Expect = 1e-04
Identities = 29/65 (44%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Frame = +2
Query: 14 SSSLPPSSSS--SSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP----SKCRHK 67
S+S PSSSS SS +S S SP SS + PSPP PS S P P S +
Sbjct: 107 SNSASPSSSSRGSSRASPSRSRSPPSSSAASSPSPPAPSSSSPSPTPSSPPSSLSPAASR 286
Query: 68 PPKPP 72
PP PP
Sbjct: 287 PPTPP 301
Score = 38.5 bits (88), Expect = 0.002
Identities = 27/65 (41%), Positives = 31/65 (47%), Gaps = 6/65 (9%)
Frame = +2
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP------SKCRH 66
S S PPSSS++SS S +P SS P PS PP S S S P P S +
Sbjct: 161 SRSRSPPSSSAASSPSPP---APSSSSPSPTPSSPPSSLSPAASRPPTPPTHSTASSSKR 331
Query: 67 KPPKP 71
PP P
Sbjct: 332 APPSP 346
Score = 28.5 bits (62), Expect = 1.7
Identities = 18/41 (43%), Positives = 20/41 (47%)
Frame = +3
Query: 34 SPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPRL 74
SP S +LP+P PPP H P P R PP P RL
Sbjct: 93 SPSHSRTLPRP-PPPLVDPHAP--PLRARALRRVPPPPLRL 206
>TC226052 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_13), partial
(39%)
Length = 941
Score = 42.4 bits (98), Expect = 1e-04
Identities = 27/68 (39%), Positives = 37/68 (53%), Gaps = 7/68 (10%)
Frame = +1
Query: 14 SSSLPPSSSSSSSSSTSLHLSPCSSH--SLPQPSPPPPSKSHFPS-----LPRFPSKCRH 66
+SS PS SSS S++ PC+S S P+ +PPPPS + P+ PR + R
Sbjct: 433 TSSASPSPSSSVSAAAPSPPPPCTSSGPSSPRATPPPPSTTSPPTKTRSRAPRS*ATSRF 612
Query: 67 KPPKPPRL 74
PP+PP L
Sbjct: 613 LPPRPPLL 636
Score = 41.6 bits (96), Expect = 2e-04
Identities = 30/82 (36%), Positives = 40/82 (48%), Gaps = 18/82 (21%)
Frame = +1
Query: 18 PPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRF----------PSKCRHK 67
PPS+++ +SS S SP SS S PSPPPP S PS PR P+K R +
Sbjct: 409 PPSATAPRTSSASP--SPSSSVSAAAPSPPPPCTSSGPSSPRATPPPPSTTSPPTKTRSR 582
Query: 68 PPK--------PPRLFMIIDPI 81
P+ PPR ++ P+
Sbjct: 583 APRS*ATSRFLPPRPPLLSPPL 648
Score = 31.2 bits (69), Expect = 0.26
Identities = 24/82 (29%), Positives = 33/82 (39%), Gaps = 19/82 (23%)
Frame = +1
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCS-SHSLPQPSP------------------PPPSK 51
P + + PP ++SSS + LSP + +H+LP SP P PS
Sbjct: 283 PTNPPANPPPPTTSSSPTRLTSLSPSA*THALP*GSPPRTISPPSATAPRTSSASPSPSS 462
Query: 52 SHFPSLPRFPSKCRHKPPKPPR 73
S + P P C P PR
Sbjct: 463 SVSAAAPSPPPPCTSSGPSSPR 528
Score = 29.6 bits (65), Expect = 0.76
Identities = 24/67 (35%), Positives = 32/67 (46%), Gaps = 4/67 (5%)
Frame = +1
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPS--LPRFPSKCRHKP 68
P S++L P+ S S S LS +S + P +PPPP+ S P+ PS H
Sbjct: 205 PTRSATLTPTPSPPSPRSDPSPLS--TSPTNPPANPPPPTTSSSPTRLTSLSPSA*THAL 378
Query: 69 P--KPPR 73
P PPR
Sbjct: 379 P*GSPPR 399
>TC226524 similar to UP|Q8VYJ5 (Q8VYJ5) At1g68530/T26J14_10, partial (39%)
Length = 608
Score = 42.0 bits (97), Expect = 1e-04
Identities = 22/48 (45%), Positives = 29/48 (59%)
Frame = +1
Query: 3 TLSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPS 50
TLS S+ P SSSS PPS+S + + ++ +P SSH P SP PS
Sbjct: 193 TLSKSSAPPSSSSSSPPSTSCPNPAPSTSSTTPASSHPSPAASPSQPS 336
Score = 32.7 bits (73), Expect = 0.090
Identities = 22/48 (45%), Positives = 29/48 (59%), Gaps = 1/48 (2%)
Frame = +1
Query: 12 LSSSSLPP-SSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLP 58
LS SS PP SSSSS S++ + +P +S + P S P P+ S PS P
Sbjct: 196 LSKSSAPPSSSSSSPPSTSCPNPAPSTSSTTPASSHPSPAAS--PSQP 333
>AI736821 weakly similar to GP|11036868|gb| PxORF73 peptide {Plutella
xylostella granulovirus}, partial (29%)
Length = 430
Score = 42.0 bits (97), Expect = 1e-04
Identities = 27/74 (36%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Frame = +2
Query: 5 SYSTLHPLSSSS-----LPPSSSSSSSSSTSLHLSP-CSSHSLPQPSPPPPSKSHFPSLP 58
S S LHPL+ S L + S + S P C LP P+PPPP P+ P
Sbjct: 122 STSLLHPLNDSIFQWILLSSLTPHSPPLTLSARFGPECPISPLPSPTPPPPITPPSPTSP 301
Query: 59 RFPSKCRHKPPKPP 72
P++ R PP PP
Sbjct: 302 PPPARERTAPPPPP 343
Score = 33.1 bits (74), Expect = 0.069
Identities = 24/76 (31%), Positives = 32/76 (41%)
Frame = +2
Query: 9 LHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKP 68
+ PL S + PP + S +S P + P P PPPP P + R P
Sbjct: 239 ISPLPSPTPPPPITPPSPTSPP---PPARERTAPPPPPPPPP-------PTRTTAPRSSP 388
Query: 69 PKPPRLFMIIDPILLL 84
P PP L PI++L
Sbjct: 389 PPPPPL--PTSPIMIL 430
>TC226484 similar to GB|AAP21355.1|30102874|BT006547 At1g75690 {Arabidopsis
thaliana;} , partial (50%)
Length = 427
Score = 41.6 bits (96), Expect = 2e-04
Identities = 22/58 (37%), Positives = 28/58 (47%), Gaps = 6/58 (10%)
Frame = +1
Query: 123 PCERCAGNGGMKCVFC-NDGKMKQEMG-----LIDCKVCKGSGLILCKKCGGSGYSRR 174
PC C G+G KC FC +G + E+G + C C G G + C C GSG R
Sbjct: 25 PCFPCNGSGAQKCRFCLGNGNVTVELGGGEEEVSRCINCDGVGSLTCTTCQGSGIQPR 198
>TC226486 similar to GB|AAP21355.1|30102874|BT006547 At1g75690 {Arabidopsis
thaliana;} , partial (76%)
Length = 952
Score = 41.2 bits (95), Expect = 3e-04
Identities = 31/98 (31%), Positives = 41/98 (41%), Gaps = 17/98 (17%)
Frame = +1
Query: 94 LDTQTLLAT----VSVLAAIALSLFLGLKGDPV-------PCERCAGNGGMKCVFC-NDG 141
LD T++A VSV I + +F + D PC C G+G KC FC G
Sbjct: 268 LDQNTVVAISVGLVSVAVGIGIPVFYETQIDNAAKRDNTQPCFPCNGSGSQKCRFCLGSG 447
Query: 142 KMKQEMG-----LIDCKVCKGSGLILCKKCGGSGYSRR 174
+ E+G + C C G + C C GSG R
Sbjct: 448 NVTVELGGGEKEVSRCINCDAVGSLTCTTCQGSGIQPR 561
>TC205979 homologue to UP|Q6XPS8 (Q6XPS8) Drought-induced protein 1, complete
Length = 638
Score = 41.2 bits (95), Expect = 3e-04
Identities = 20/52 (38%), Positives = 26/52 (49%)
Frame = +1
Query: 124 CERCAGNGGMKCVFCNDGKMKQEMGLIDCKVCKGSGLILCKKCGGSGYSRRL 175
C C G G + C+ C+ + G + C C GSG + C CGGSG R L
Sbjct: 187 CPTCNGTGRVTCL-CS----RWSDGDVGCSTCSGSGRMACSSCGGSGTGRPL 327
>CO985879
Length = 695
Score = 41.2 bits (95), Expect = 3e-04
Identities = 26/59 (44%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Frame = -1
Query: 16 SLPPSSSSSSSSSTSLHLSPCS-SHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPKPPR 73
S PP SSS S+S T SP + S P PPP S + PS P PS + PK PR
Sbjct: 548 SPPPRSSSPSASPTPTAASPSRPAPSPPSWRPPPTSTAPPPSAPSLPSSSCARSPKAPR 372
Score = 33.9 bits (76), Expect = 0.040
Identities = 22/61 (36%), Positives = 27/61 (44%)
Frame = -1
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKPPK 70
P +S+ PP S+ S SS+ S P PSP P S P PR P R P +
Sbjct: 455 PPPTSTAPPPSAPSLPSSSCARSPKAPRSSAPTPSPSP---SWSP*WPRPPQGARSTPSE 285
Query: 71 P 71
P
Sbjct: 284 P 282
>TC209731 similar to UP|Q8W3M8 (Q8W3M8) 6b-interacting protein 1, partial
(20%)
Length = 581
Score = 41.2 bits (95), Expect = 3e-04
Identities = 31/70 (44%), Positives = 34/70 (48%)
Frame = +1
Query: 5 SYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKC 64
S S P +S+S PP S TSL SP SS SLP P P PP +S L P
Sbjct: 379 SVSPNPPSASASAPPLSHKLPQ--TSLSPSPTSS-SLPSPYPLPPLRSLSLPLHILPPPP 549
Query: 65 RHKPPKPPRL 74
PP PPRL
Sbjct: 550 PPNPPPPPRL 579
>TC205978 homologue to UP|Q6XPS8 (Q6XPS8) Drought-induced protein 1, complete
Length = 645
Score = 41.2 bits (95), Expect = 3e-04
Identities = 20/52 (38%), Positives = 26/52 (49%)
Frame = +3
Query: 124 CERCAGNGGMKCVFCNDGKMKQEMGLIDCKVCKGSGLILCKKCGGSGYSRRL 175
C C G G + C+ C+ + G + C C GSG + C CGGSG R L
Sbjct: 246 CPTCNGTGRVTCL-CS----RWSDGDVGCSTCSGSGRMACSSCGGSGTGRPL 386
>TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase , complete
Length = 1675
Score = 40.8 bits (94), Expect = 3e-04
Identities = 27/73 (36%), Positives = 37/73 (49%), Gaps = 1/73 (1%)
Frame = +1
Query: 2 ATLSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFP 61
AT + + P S + P SSSSS+ + TS + CS+ S P PS P P + P+ P P
Sbjct: 295 ATWEWPSRRPSSLRTSPTSSSSSTPTPTS-SAARCSTSSTPPPSSPAPRSTPPPTPPSPP 471
Query: 62 S-KCRHKPPKPPR 73
+ PP P R
Sbjct: 472 APTSASSPPAPAR 510
Score = 28.9 bits (63), Expect = 1.3
Identities = 25/67 (37%), Positives = 33/67 (48%), Gaps = 5/67 (7%)
Frame = +1
Query: 11 PLSSSSLP-PSSSSSSSSSTSLH-LSPCSSHSLPQPSPPPPS--KSHFPS-LPRFPSKCR 65
P S+SS P P+ S +S +STS SP S+ P S PP+ S FP+ P+
Sbjct: 475 PTSASSPPAPARSPASHASTSSRGTSPSSAPLFPLSSVTPPTQLSSSFPTPSTSSPTSRG 654
Query: 66 HKPPKPP 72
P PP
Sbjct: 655 SSPASPP 675
Score = 28.5 bits (62), Expect = 1.7
Identities = 27/72 (37%), Positives = 34/72 (46%), Gaps = 10/72 (13%)
Frame = +1
Query: 5 SYSTLHPLSS--------SSLPPSSSSSSSSSTSLHLSPCSSHSLPQPS-PPPPSKSHFP 55
S + L PLSS SS P S+SS +S S SP ++ S P P+ P S S P
Sbjct: 556 SSAPLFPLSSVTPPTQLSSSFPTPSTSSPTSRGSSPASPPTASSAPAPTWTLPASVSSSP 735
Query: 56 -SLPRFPSKCRH 66
+L P RH
Sbjct: 736 ITLTSTPRTYRH 771
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 40.8 bits (94), Expect = 3e-04
Identities = 29/71 (40%), Positives = 37/71 (51%), Gaps = 4/71 (5%)
Frame = +3
Query: 6 YSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLP--QPSPPPPSKSHFPSLPRFPSK 63
+ T P S++PPS++ S S+TS SP +S + P PSP PPS PS P PS
Sbjct: 222 HRTPAPSPPSTMPPSATPPSPSATS---SPSASPTAPPTSPSPSPPSSPSPPSAPS-PSS 389
Query: 64 CRHKPP--KPP 72
PP PP
Sbjct: 390 TAPSPPSASPP 422
Score = 34.7 bits (78), Expect = 0.024
Identities = 24/69 (34%), Positives = 30/69 (42%), Gaps = 4/69 (5%)
Frame = +3
Query: 8 TLHPLSSSSLPPSS----SSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSK 63
T P S S PPSS S+ S SST+ S + P PSPP P+ + + P S
Sbjct: 315 TAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSP 494
Query: 64 CRHKPPKPP 72
P P
Sbjct: 495 TSRTSPTSP 521
Score = 34.7 bits (78), Expect = 0.024
Identities = 24/63 (38%), Positives = 30/63 (47%), Gaps = 2/63 (3%)
Frame = +3
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFP--SLPRFPSKCRHKP 68
P S S PPS +S++S+++ SP S S PSP SK P S P PS
Sbjct: 420 PTSPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPS 599
Query: 69 PKP 71
P P
Sbjct: 600 PTP 608
Score = 30.8 bits (68), Expect = 0.34
Identities = 24/78 (30%), Positives = 36/78 (45%)
Frame = +3
Query: 5 SYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKC 64
S +T P S +S+ P ++S ++ S SS + P P P K PS+ F SK
Sbjct: 624 SPATSTPTSHTSISPITASKATFLLP*LCSTASSF*I-SPLTPSPEKYLLPSVT*FHSKT 800
Query: 65 RHKPPKPPRLFMIIDPIL 82
PP P + +I +L
Sbjct: 801 SPSPPTPSPVPFLIPYLL 854
Score = 28.1 bits (61), Expect = 2.2
Identities = 16/45 (35%), Positives = 24/45 (52%)
Frame = +3
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSL 57
+S+S PP+ SSSS + S SP + P PSP + + S+
Sbjct: 528 TSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSI 662
Score = 26.2 bits (56), Expect = 8.4
Identities = 15/43 (34%), Positives = 19/43 (43%)
Frame = +3
Query: 14 SSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPS 56
+S PS +S+S SP S P PSP P S P+
Sbjct: 504 TSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPA 632
>TC219140 similar to GB|AAO44047.1|28466877|BT004781 At2g39020 {Arabidopsis
thaliana;} , partial (75%)
Length = 730
Score = 40.8 bits (94), Expect = 3e-04
Identities = 28/63 (44%), Positives = 31/63 (48%), Gaps = 1/63 (1%)
Frame = +3
Query: 11 PLSSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRH-KPP 69
P S+S P +S+SSST S S S P PSPP P S P P PS KPP
Sbjct: 90 PASASPPLPMFPTSTSSSTRWPSSNASPTSSPPPSPPSPPPSSLPH-PSNPSPSSF*KPP 266
Query: 70 KPP 72
PP
Sbjct: 267 PPP 275
Score = 27.3 bits (59), Expect = 3.8
Identities = 16/38 (42%), Positives = 19/38 (49%)
Frame = +3
Query: 13 SSSSLPPSSSSSSSSSTSLHLSPCSSHSLPQPSPPPPS 50
S +S PP S S S+ H S S S +P PPP S
Sbjct: 168 SPTSSPPPSPPSPPPSSLPHPSNPSPSSF*KPPPPPSS 281
>CO982258
Length = 732
Score = 40.4 bits (93), Expect = 4e-04
Identities = 38/121 (31%), Positives = 54/121 (44%), Gaps = 7/121 (5%)
Frame = -1
Query: 3 TLSYSTLHPLSSSSLPPSSSSSSSSSTSLHLSPCS--SHSLPQPSPPPPSKSHFPSLPRF 60
T + +P + S + PSSSSSSS++T SP + + S P PPP + PS+P
Sbjct: 654 TATLVNTNPSTGSCIFPSSSSSSSTTTMPKSSPPTPPTQSSPIALPPPSPLTPEPSIPTA 475
Query: 61 PSKCRHKPPKPPRLFMIIDPILLLNGFGGA-----GFYLDTQTLLATVSVLAAIALSLFL 115
P P F P LN A GF D+ ++ T SV ++ L F+
Sbjct: 474 PPTSSSSGTGP---FGYGAPPSELNSSNPASGIMPGFASDSPPVVNTTSVSRSVGLRPFV 304
Query: 116 G 116
G
Sbjct: 303 G 301
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,127,700
Number of Sequences: 63676
Number of extensions: 386570
Number of successful extensions: 18567
Number of sequences better than 10.0: 2106
Number of HSP's better than 10.0 without gapping: 8739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 13733
length of query: 175
length of database: 12,639,632
effective HSP length: 91
effective length of query: 84
effective length of database: 6,845,116
effective search space: 574989744
effective search space used: 574989744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0546.10


