

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
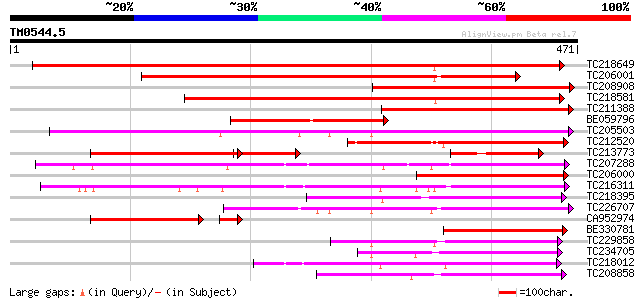
Query= TM0544.5
(471 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 414 e-116
TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 371 e-103
TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-... 298 4e-81
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 282 2e-76
TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like pr... 278 4e-75
BE059796 187 7e-48
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 186 1e-47
TC212520 186 3e-47
TC213773 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 134 8e-43
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 171 8e-43
TC206000 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like pr... 166 2e-41
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 147 1e-35
TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 125 4e-29
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 125 5e-29
CA952974 similar to GP|17381265|gb| AT5g18840/F17K4_90 {Arabidop... 112 3e-27
BE330781 weakly similar to GP|17381265|gb AT5g18840/F17K4_90 {Ar... 117 8e-27
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 116 2e-26
TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solu... 110 1e-24
TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 108 5e-24
TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%) 104 1e-22
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 414 bits (1065), Expect = e-116
Identities = 212/447 (47%), Positives = 302/447 (67%), Gaps = 5/447 (1%)
Frame = +2
Query: 20 QQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGA 79
Q R + ++ L+ LG FG GY+SPTQSAI+ DL L+V++FS+FGS+ +GA
Sbjct: 146 QAIRDSSISVFACVLIVALGPIQFGFTAGYTSPTQSAIINDLGLSVSEFSLFGSLSNVGA 325
Query: 80 MIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYV 139
M+GAI SG++A+Y GR+ ++ + + I+GWL I+F+K + +LY+GRLL G G+G++SY
Sbjct: 326 MVGAIASGQIAEYIGRKGSLMIASIPNIIGWLAISFAKDSSFLYMGRLLEGFGVGIISYT 505
Query: 140 VPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLP 199
VPVY+AEI+P NLRGG +++QL + G+ L YL+G FV WRILA+IG +PC + +L
Sbjct: 506 VPVYIAEISPPNLRGGLVSVNQLSVTIGIMLAYLLGIFVEWRILAIIGILPCTILIPALF 685
Query: 200 FIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQW 259
FIP+SPRWLAK+G +E +++LQ LRG + D+ E EI+ + + L Q
Sbjct: 686 FIPESPRWLAKMGMTEEFETSLQVLRGFDTDISVEVNEIKRAVASTNTRITVRFADLKQR 865
Query: 260 KYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES-IGTIAMVAVKIPVTVLGVL 318
+Y L IG+GL+ILQQ GING++FY+++IF +AG S S T + AV++ T L +
Sbjct: 866 RYWLPLMIGIGLLILQQLSGINGVLFYSSTIFRNAGISSSDAATFGVGAVQVLATSLTLW 1045
Query: 319 LMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQG----LHEWKGVSPILALVGVLVYVGSY 374
L DKSGRR LL++SA G + A+ F ++ G+ L+LVGV+ V ++
Sbjct: 1046LADKSGRRLLLIVSATGMSFSLLVVAITFYIKASISETSSLYGILSTLSLVGVVAMVIAF 1225
Query: 375 SLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSS 434
SLGMGA+PW+IMSEI PIN+KG AGS+ T NWL SW+V+ N L+ WSS GTF I++
Sbjct: 1226SLGMGAMPWIIMSEILPINIKGLAGSVATLANWLFSWLVTLTANMLLDWSSGGTFTIYAV 1405
Query: 435 ICGFTVLFVAKLVPETKGRTLEEIQAS 461
+C TV+FV VPETKG+T+EEIQ S
Sbjct: 1406VCALTVVFVTIWVPETKGKTIEEIQWS 1486
>TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (66%)
Length = 951
Score = 371 bits (953), Expect = e-103
Identities = 187/319 (58%), Positives = 236/319 (73%), Gaps = 4/319 (1%)
Frame = +3
Query: 110 WLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMS 169
WL + FSK ++ L +GR G GIG++SYVVPVY+AEI PKNLRGG T +QL+I G S
Sbjct: 3 WLAVFFSKGSYSLDMGRFFTGYGIGVISYVVPVYIAEIAPKNLRGGLATTNQLLIVTGGS 182
Query: 170 LTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNA 229
+++L+G+ +NWR LAL G +PC L+ L FIP+SPRWLAKVG KE AL RLRGK+A
Sbjct: 183 VSFLLGSVINWRELALAGLVPCICLLVGLCFIPESPRWLAKVGREKEFQLALSRLRGKHA 362
Query: 230 DVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANS 289
D+ EAAEI DY ETL+ + ++ LFQ KY+ S+ IGVGLM QQ GINGI FY
Sbjct: 363 DISDEAAEILDYIETLESLPKTKLLDLFQSKYVHSVVIGVGLMACQQSVGINGIGFYTAE 542
Query: 290 IFVSAGFSE-SIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFL 348
IFV+AG S GTIA ++IP T+LG +LMDKSGRRPL+++SAAGT LGCF+AA AF
Sbjct: 543 IFVAAGLSSGKAGTIAYACIQIPFTLLGAILMDKSGRRPLVMVSAAGTFLGCFVAAFAFF 722
Query: 349 LQG---LHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFF 405
L+ L EW PILA GVL+Y+ ++S+G+G++PWVIMSEIFPI++KG+AGSLV
Sbjct: 723 LKDQSLLPEW---VPILAFAGVLIYIAAFSIGLGSVPWVIMSEIFPIHLKGTAGSLVVLV 893
Query: 406 NWLCSWIVSYAFNFLMSWS 424
WL +W+VSY FNFLMSWS
Sbjct: 894 AWLGAWVVSYTFNFLMSWS 950
>TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like
protein, partial (28%)
Length = 753
Score = 298 bits (763), Expect = 4e-81
Identities = 145/168 (86%), Positives = 160/168 (94%)
Frame = +2
Query: 302 TIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPI 361
TIA+VAVKIP+T +GVLLMDKSGRRPLLL+SA GTC+GCFLAAL+F+LQ LH+WKGVSPI
Sbjct: 2 TIAIVAVKIPMTTIGVLLMDKSGRRPLLLVSAVGTCVGCFLAALSFVLQDLHKWKGVSPI 181
Query: 362 LALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLM 421
LALVGVLVYVGSYS+GMGAIPWVIMSEIFPINVKGSAGSLVT +WLCSWI+SY+FNFLM
Sbjct: 182 LALVGVLVYVGSYSIGMGAIPWVIMSEIFPINVKGSAGSLVTLVSWLCSWIISYSFNFLM 361
Query: 422 SWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTKR 469
SWSS GTF +FSSICGFTVLFVAKLVPETKGRTLEEIQASL+S S+KR
Sbjct: 362 SWSSAGTFLMFSSICGFTVLFVAKLVPETKGRTLEEIQASLNSFSSKR 505
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 282 bits (722), Expect = 2e-76
Identities = 143/321 (44%), Positives = 212/321 (65%), Gaps = 5/321 (1%)
Frame = +2
Query: 146 EITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSP 205
EI P +L GG +++QL I G+ L YL+G FVNWRILA++G +PC + L FIP+SP
Sbjct: 5 EIAPNHLIGGLGSVNQLSITIGIMLAYLLGLFVNWRILAILGILPCTVLIPGLFFIPESP 184
Query: 206 RWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSL 265
RWLAK+G + E +++LQ LRG + D+ E EI+ + ++ L + +Y L
Sbjct: 185 RWLAKMGMIDEFETSLQVLRGFDTDISVEVHEIKRSVASTGKRAAIRFADLKRKRYWFPL 364
Query: 266 TIGVGLMILQQFGGINGIVFYANSIFVSAGFSES-IGTIAMVAVKIPVTVLGVLLMDKSG 324
+G+GL++LQQ GINGI+FY+ +IF +AG S S T+ + AV++ T + L+DKSG
Sbjct: 365 MVGIGLLVLQQLSGINGILFYSTTIFANAGISSSEAATVGLGAVQVIATGISTWLVDKSG 544
Query: 325 RRPLLLLSAAGTCLGCFLAALAFLLQGL----HEWKGVSPILALVGVLVYVGSYSLGMGA 380
RR LL++S++ + + ++AF L+G+ + I+++VG++ V +SLG+G
Sbjct: 545 RRLLLIISSSVMTVSLLIVSIAFYLEGVVSEDSHLFSILGIVSIVGLVAMVIGFSLGLGP 724
Query: 381 IPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTV 440
IPW+IMSEI P+N+KG AGS+ T NWL SW ++ N L++WSS GTF I++ + FT+
Sbjct: 725 IPWLIMSEILPVNIKGLAGSIATMGNWLISWGITMTANLLLNWSSGGTFTIYTVVAAFTI 904
Query: 441 LFVAKLVPETKGRTLEEIQAS 461
F+A VPETKGRTLEEIQ S
Sbjct: 905 AFIAMWVPETKGRTLEEIQFS 967
>TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like protein,
partial (27%)
Length = 786
Score = 278 bits (711), Expect = 4e-75
Identities = 135/159 (84%), Positives = 146/159 (90%)
Frame = +3
Query: 310 IPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLV 369
IP+T LGVLLMDKSGRRPLLL+SA+GTCLGCFLAAL+F LQ LH+WK SPILAL GVLV
Sbjct: 87 IPMTALGVLLMDKSGRRPLLLISASGTCLGCFLAALSFTLQDLHKWKEGSPILALAGVLV 266
Query: 370 YVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTF 429
Y GS+SLGMG IPWVIMSEIFP NVKGSAGSLVT +WLCSWIVSYAFNFLMSWSS GTF
Sbjct: 267 YTGSFSLGMGGIPWVIMSEIFPTNVKGSAGSLVTLVSWLCSWIVSYAFNFLMSWSSAGTF 446
Query: 430 FIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTK 468
FIFSSICGFT+LFVAKLVPETKGRTLEE+QASL+ +STK
Sbjct: 447 FIFSSICGFTILFVAKLVPETKGRTLEEVQASLNPYSTK 563
>BE059796
Length = 410
Score = 187 bits (476), Expect = 7e-48
Identities = 95/131 (72%), Positives = 110/131 (83%)
Frame = +1
Query: 184 ALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTE 243
AL+G IPC QLL L FIP+SPRWLAK GH + S+S LQRLRGKNADV QEA EIRD+TE
Sbjct: 1 ALLGIIPCIVQLLGLFFIPESPRWLAKFGHWERSESVLQRLRGKNADVSQEATEIRDFTE 180
Query: 244 TLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTI 303
LQR+TE SIIGLFQ +Y+ SLT+GVGLMILQQFGG+NGI FYA+SIF+SAGFS SIG I
Sbjct: 181 ALQRETE-SIIGLFQLQYLKSLTVGVGLMILQQFGGVNGIAFYASSIFISAGFSGSIGMI 357
Query: 304 AMVAVKIPVTV 314
MVAV++ T+
Sbjct: 358 TMVAVQVLHTI 390
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 186 bits (473), Expect = 1e-47
Identities = 128/463 (27%), Positives = 219/463 (46%), Gaps = 28/463 (6%)
Frame = +1
Query: 34 LVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVADYA 93
++A + S G IG S I DL ++ Q I I+ + ++IG+ ++G+ +D+
Sbjct: 271 MLASMTSILLGYDIGVMSGAAIYIKRDLKVSDEQIEILLGIINLYSLIGSCLAGRTSDWI 450
Query: 94 GRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLR 153
GRR +G ++G ++ F +L GR + G GIG + PVY AE++P + R
Sbjct: 451 GRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYTAEVSPASSR 630
Query: 154 GGFTTMHQLMICCGMSLTYL-------IGAFVNWRILALIGAIPCFAQLLSLPFIPDSPR 206
G T+ ++ I G+ L Y+ + V WR++ +GAIP + +P+SPR
Sbjct: 631 GFLTSFPEVFINGGILLGYISNYGFSKLTLKVGWRMMLGVGAIPSVVLTEGVLAMPESPR 810
Query: 207 WLAKVGHLKESDSALQRLRGKNADVHQEAAEIR----------DYTETLQRQTEASIIGL 256
WL G L E+ L + + AEI+ D + +Q+ +
Sbjct: 811 WLVMRGRLGEARKVLNKTSDSKEEAQLRLAEIKQAAGIPESCNDDVVQVNKQSNGEGVWK 990
Query: 257 FQWKYMTS-----LTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES----IGTIAMVA 307
+ Y T + +G+ QQ G++ +V Y+ IF AG + + T+A+
Sbjct: 991 ELFLYPTPAIRHIVIAALGIHFFQQASGVDAVVLYSPRIFEKAGITNDTHKLLATVAVGF 1170
Query: 308 VKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPI-LALVG 366
VK + +D+ GRRPLLL S G L A++ + E K + + ++
Sbjct: 1171VKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTVIDHSERKLMWAVGSSIAM 1350
Query: 367 VLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAF-NFLMSWSS 425
VL YV ++S+G G I WV SEIFP+ ++ + N S +VS F + + +
Sbjct: 1351VLAYVATFSIGAGPITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVSMTFLSLTRAITI 1530
Query: 426 TGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTK 468
G FF++ I +F ++PET+G+TLE+++ S + +K
Sbjct: 1531GGAFFLYCGIATVGWIFFYTVLPETRGKTLEDMEGSFGTFRSK 1659
>TC212520
Length = 720
Score = 186 bits (471), Expect = 3e-47
Identities = 92/186 (49%), Positives = 131/186 (69%), Gaps = 2/186 (1%)
Frame = +3
Query: 281 NGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGC 340
NGI F ++ ++ S +IGTI ++I LG L+DK+GR+PL+LLS +G GC
Sbjct: 3 NGICFIP-AVXXTSRISPTIGTITYACLQIVXXGLGAALIDKAGRKPLILLSGSGLVAGC 179
Query: 341 FLAALAFLLQGLHEWKGVS--PILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSA 398
A+AF L+ +HE GV P LA+ G+LVY+GS+S+GMGAIPWV+M EIFP+N+KG A
Sbjct: 180 TFVAVAFYLK-VHE-VGVEAVPALAVTGILVYIGSFSIGMGAIPWVVMCEIFPVNIKGLA 353
Query: 399 GSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEI 458
GS+ T NW +W+ SY FNF MSWSS GTF ++++I +LF+ VPE+KG++LE++
Sbjct: 354 GSVATLVNWFGAWLCSYTFNFFMSWSSYGTFILYAAINALDILFIIVAVPESKGKSLEQL 533
Query: 459 QASLSS 464
QA ++S
Sbjct: 534 QADINS 551
>TC213773 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (49%)
Length = 730
Score = 134 bits (338), Expect(2) = 8e-43
Identities = 65/126 (51%), Positives = 89/126 (70%)
Frame = +3
Query: 68 FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRL 127
FS+FGS++TIGAM+GAI SG++ D+ GR+ AM S FCI G + FSK ++ L +GR
Sbjct: 12 FSMFGSLVTIGAMLGAISSGRITDFIGRKGAMRISAGFCITGCFFVFFSKGSYSLDLGRF 191
Query: 128 LVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVNWRILALIG 187
G GIG++SYVVPVY+ EI PKNLR T +QL+I S+++L+G+ +NWR LAL G
Sbjct: 192 FTGYGIGVISYVVPVYIVEIAPKNLREELATTNQLLIVTEASVSFLLGSVINWRKLALAG 371
Query: 188 AIPCFA 193
+ C A
Sbjct: 372 LVSCIA 389
Score = 91.3 bits (225), Expect = 9e-19
Identities = 43/77 (55%), Positives = 54/77 (69%)
Frame = +2
Query: 367 VLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSST 426
+L+Y+ +YS+G G +PWVIMSE G AGSLV NWL +WIVSY FNFLMSWSS
Sbjct: 521 ILIYIAAYSIGEGPVPWVIMSE-------GIAGSLVVLVNWLGAWIVSYTFNFLMSWSSP 679
Query: 427 GTFFIFSSICGFTVLFV 443
GT F+++ T+LFV
Sbjct: 680 GT*FLYAGSSLLTILFV 730
Score = 57.8 bits (138), Expect(2) = 8e-43
Identities = 31/55 (56%), Positives = 36/55 (65%)
Frame = +2
Query: 187 GAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDY 241
GA L+ L FIP+SPRWLAKVG K AL+RLRGK+ D+ EAAEI Y
Sbjct: 368 GASVLHCWLVGLCFIPESPRWLAKVGREK*FQLALRRLRGKDVDISDEAAEILIY 532
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 171 bits (432), Expect = 8e-43
Identities = 132/478 (27%), Positives = 218/478 (44%), Gaps = 34/478 (7%)
Frame = +1
Query: 22 QRPFTATLTLSTLVAVLGSYAFGAAIGYSS--PTQSAIMPDLNLTVAQ------------ 67
+ FTA + +V LG FG +G S P+ + + V +
Sbjct: 82 EHKFTAYFAFTCVVGALGGSLFGYDLGVSGGVPSMDDFLKEFFPKVYRRKQMHLHETDYC 261
Query: 68 ------FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWW 121
++F S L A++ + + GR+ ++ L + G ++ +K
Sbjct: 262 KYDDQVLTLFTSSLYFSALVMTFFASFLTRKKGRKASIIVGALSFLAGAILNAAAKNIAM 441
Query: 122 LYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVN-- 179
L +GR+L+G GIG + VP+Y++E+ P RG + Q C G+ + L+ F
Sbjct: 442 LIIGRVLLGGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQFTTCAGILIANLVNYFTEKI 621
Query: 180 ----WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEA 235
WRI + +P FA L+ ++P L + G L ++ LQR+RG +V E
Sbjct: 622 HPYGWRISLGLAGLPAFAMLVGGICCAETPNSLVEQGRLDKAKQVLQRIRGTE-NVEAEF 798
Query: 236 AEIRDYTETLQRQTEASIIGLFQWKYMTSLTIG-VGLMILQQFGGINGIVFYANSIFVSA 294
++++ +E Q ++ L + KY L IG +G+ QQ G N I+FYA IF S
Sbjct: 799 EDLKEASEEAQA-VKSPFRTLLKRKYRPQLIIGALGIPAFQQLTGNNSILFYAPVIFQSL 975
Query: 295 GFSESIGTIAMVAVK---IPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLL-- 349
GF + + + TV+ + L+DK GRR L A + C + A L
Sbjct: 976 GFGANASLFSSFITNGALLVATVISMFLVDKYGRRKFFL-EAGFEMICCMIITGAVLAVN 1152
Query: 350 --QGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNW 407
G KGVS L +V + ++V +Y G + W++ SE+FP+ ++ SA S+V N
Sbjct: 1153FGHGKEIGKGVSAFLVVV-IFLFVLAYGRSWGPLGWLVPSELFPLEIRSSAQSIVVCVNM 1329
Query: 408 LCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSH 465
+ + +V+ F + G F +F+S+ F FV L+PETK +EEI +H
Sbjct: 1330IFTALVAQLFLMSLCHLKFGIFLLFASLIIFMSFFVFFLLPETKKVPIEEIYLLFENH 1503
>TC206000 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like protein,
partial (22%)
Length = 602
Score = 166 bits (420), Expect = 2e-41
Identities = 75/126 (59%), Positives = 100/126 (78%)
Frame = +1
Query: 339 GCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSA 398
GC++A +AF ++ + W PILA GVL+Y+ ++S+G+G++PWVIMSEIFPI++KG+A
Sbjct: 1 GCWMAGIAFFVRHHNSWLEWVPILAFAGVLIYIAAFSIGLGSVPWVIMSEIFPIHLKGTA 180
Query: 399 GSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEI 458
GSLV WL +W+VSY FNFLMSWSS GT F+++ T+LFVAKLVPETKG+TLEEI
Sbjct: 181 GSLVVLVAWLGAWVVSYTFNFLMSWSSPGTLFLYAGCSLLTILFVAKLVPETKGKTLEEI 360
Query: 459 QASLSS 464
QA +SS
Sbjct: 361 QACISS 378
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 147 bits (371), Expect = 1e-35
Identities = 132/481 (27%), Positives = 222/481 (45%), Gaps = 41/481 (8%)
Frame = +1
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSA------IMPDL------NLTVAQF----- 68
T +T++ +VA +G FG IG S S P + + TV Q+
Sbjct: 154 TPFVTVTCIVAAMGGLIFGYDIGISGGVTSMDPFLLKFFPSVFRKKNSDKTVNQYCQYDS 333
Query: 69 ---SIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVG 125
++F S L + A++ ++V+ V GR+++M F L ++G L+ F++ W L VG
Sbjct: 334 QTLTMFTSSLYLAALLSSLVASTVTRRFGRKLSMLFGGLLFLVGALINGFAQHVWMLIVG 513
Query: 126 RLLVGCGIGLLSYV---VPVYVAEITPKNLRG--GFTTMHQLMICCGMSLTYLIG----A 176
R+L+G GIG + V +P+ I G F Q I G + L+
Sbjct: 514 RILLGFGIGFANQVCATLPI*NGFIQI*RSIGTLAFNCQSQFGIPRGQCVELLLWLKSMV 693
Query: 177 FVNWRILALIGA-IPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEA 235
W+I GA +P + +PD+P + + G +++ + LQR+RG + +V +E
Sbjct: 694 AWGWKIEVWEGAMVPALIITVGSLVLPDTPNSMIERGDREKAKAQLQRIRGID-NVDEEF 870
Query: 236 AEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAG 295
++ +E+ Q E L Q KY LT+ V + QQ GIN I+FYA +F S G
Sbjct: 871 NDLVAASES-SSQVEHPWRNLLQRKYRPHLTMAVLIPFFQQLTGINVIMFYAPVLFSSIG 1047
Query: 296 FSESIGTIAMV---AVKIPVTVLGVLLMDKSGRRPLLLLSAAGT--CLGCFLAALA--FL 348
F + ++ V V + T + + +DK GRR L L C AA+ F
Sbjct: 1048FKDDAALMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGVQMLICQAVVAAAIGAKFG 1227
Query: 349 LQG----LHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTF 404
G L +W + +L + +YV +++ G + W++ SEIFP+ ++ +A S+
Sbjct: 1228TDGNPGDLPKWYAIVVVLF---ICIYVSAFAWSWGPLGWLVPSEIFPLEIRSAAQSINVS 1398
Query: 405 FNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSS 464
N L +++++ F ++ G F F+ FV +PETKG +EE+ +
Sbjct: 1399VNMLFTFLIAQVFLTMLCHMKFGLFLFFAFFVLIMTFFVYFFLPETKGIPIEEMGQVWQA 1578
Query: 465 H 465
H
Sbjct: 1579H 1581
>TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (40%)
Length = 1013
Score = 125 bits (314), Expect = 4e-29
Identities = 71/221 (32%), Positives = 125/221 (56%), Gaps = 5/221 (2%)
Frame = +1
Query: 247 RQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMV 306
+++E + + +FQ + + IG GL++ QQ G +++YA I SAGFS + +
Sbjct: 70 QESEGNFLEVFQGPNLKAFIIGGGLVLFQQITGQPSVLYYAGPILQSAGFSAASDATKVS 249
Query: 307 AV----KIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPIL 362
V K+ +T + VL +D GRRPLL+ +G L L L +++ G P++
Sbjct: 250 VVIGLFKLLMTWIAVLKVDDLGRRPLLIGGVSGIALSLVL------LSAYYKFLGGFPLV 411
Query: 363 ALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMS 422
A+ +L+YVG Y + G I W+++SE+FP+ +G SL N+ + +V++AF+ L
Sbjct: 412 AVGALLLYVGCYQISFGPISWLMVSEVFPLRTRGKGISLAVLTNFASNAVVTFAFSPLKE 591
Query: 423 W-SSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASL 462
+ + F +F +I ++LF+ VPETKG +LE+I++ +
Sbjct: 592 FLGAENLFLLFGAIATLSLLFIIFSVPETKGMSLEDIESKI 714
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 125 bits (313), Expect = 5e-29
Identities = 86/316 (27%), Positives = 150/316 (47%), Gaps = 25/316 (7%)
Frame = +3
Query: 178 VNWRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAE 237
+ WR++ +GAIP +++ +P+SPRWL G L E+ L ++ + A+
Sbjct: 54 LGWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAKRVLYKISESEEEARLRLAD 233
Query: 238 IRDYTETLQRQTEASII-------GLFQWKYMTS---------LTIGVGLMILQQFGGIN 281
I+D T + + + ++ G W+ + +G+ Q GI+
Sbjct: 234 IKD-TAGIPQDCDDDVVLVSKQTHGHGVWRELFLHPTPAVRHIFIASLGIHFFAQATGID 410
Query: 282 GIVFYANSIFVSAGFSES----IGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTC 337
+V Y+ IF AG + T+A+ VK ++ +D++GRR LLL S +G
Sbjct: 411 AVVLYSPRIFEKAGIKSDNYRLLATVAVGFVKTVSILVATFFLDRAGRRVLLLCSVSGLI 590
Query: 338 LGCFLAALAFLL----QGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPIN 393
L L+ + Q W + L++ VL YV ++S+G G I WV SEIFP+
Sbjct: 591 LSLLTLGLSLTVVDHSQTTLNW---AVGLSIAAVLSYVATFSIGSGPITWVYSSEIFPLR 761
Query: 394 VKGSAGSLVTFFNWLCSWIVSYAFNFLM-SWSSTGTFFIFSSICGFTVLFVAKLVPETKG 452
++ ++ N + S +++ F L + + G FF+F+ + +F L+PET+G
Sbjct: 762 LRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFFLFAGVAAVAWIFHYTLLPETRG 941
Query: 453 RTLEEIQASLSSHSTK 468
+TLEEI+ S + K
Sbjct: 942 KTLEEIEKSFGNFCRK 989
>CA952974 similar to GP|17381265|gb| AT5g18840/F17K4_90 {Arabidopsis
thaliana}, partial (19%)
Length = 372
Score = 112 bits (281), Expect(2) = 3e-27
Identities = 54/94 (57%), Positives = 70/94 (74%)
Frame = +3
Query: 68 FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRL 127
FS+FGS++TIGAM+GAI SG++ D+ GR+ AM S FCI GWLV+ FSK ++ L +GR
Sbjct: 12 FSMFGSLVTIGAMLGAISSGRITDFIGRKGAMRISAGFCITGWLVVFFSKGSYSLDLGRF 191
Query: 128 LVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQ 161
G GIG++SYVVPVY+ EI PKNLR T +Q
Sbjct: 192 FTGYGIGVISYVVPVYIVEIAPKNLREELATTNQ 293
Score = 27.3 bits (59), Expect(2) = 3e-27
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = +2
Query: 175 GAFVNWRILALIGAIPCFA 193
G+ +NWR LAL G + C A
Sbjct: 293 GSVINWRKLALAGLVSCIA 349
>BE330781 weakly similar to GP|17381265|gb AT5g18840/F17K4_90 {Arabidopsis
thaliana}, partial (20%)
Length = 364
Score = 117 bits (294), Expect = 8e-27
Identities = 60/103 (58%), Positives = 76/103 (73%)
Frame = -1
Query: 361 ILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFL 420
I A+ L+ + +YS+G+ + VIMSEIFPI+VKG AGSLV NW+ + IVSY FN L
Sbjct: 310 IFAVNDDLINIAAYSIGVAPVLDVIMSEIFPIHVKGIAGSLVVLANWIAACIVSYTFNSL 131
Query: 421 MSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLS 463
MSWSSTGT F+++ T+LFV KLVPETKG+TL EIQA +S
Sbjct: 130 MSWSSTGTLFLYAGSSLLTILFVTKLVPETKGKTL*EIQAWIS 2
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 116 bits (291), Expect = 2e-26
Identities = 69/203 (33%), Positives = 107/203 (51%), Gaps = 10/203 (4%)
Frame = +3
Query: 267 IGVGLMILQQFGGINGIVFYANSIFVSAGFSES----IGTIAMVAVKIPVTVLGVLLMDK 322
+G GL+ QQF GIN +++Y+ +I AGF + + ++ + + T+LG+ L+D
Sbjct: 24 VGAGLLAFQQFTGINTVMYYSPTIVQMAGFHANELALLLSLIVAGMNAAGTILGIYLIDH 203
Query: 323 SGRRPLLLLSAAGTCLGCFLAALAFLLQG-----LHEWKGVSPILALVGVLVYVGSYSLG 377
+GR+ L L S G + + A AF Q L+ W LA+VG+ +Y+G +S G
Sbjct: 204 AGRKKLALSSLGGVIVSLVILAFAFYKQSSTSNELYGW------LAVVGLALYIGFFSPG 365
Query: 378 MGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTG-TFFIFSSIC 436
MG +PW + SEI+P +G G + W+ + IVS F + G TF I I
Sbjct: 366 MGPVPWTLSSEIYPEEYRGICGGMSATVCWVSNLIVSETFLSIAEGIGIGSTFLIIGVIA 545
Query: 437 GFTVLFVAKLVPETKGRTLEEIQ 459
+FV VPETKG T +E++
Sbjct: 546 VVAFVFVLVYVPETKGLTFDEVE 614
>TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solute carrier
family 2 (facilitated glucose transporter), member 12
{Mus musculus;} , partial (5%)
Length = 992
Score = 110 bits (276), Expect = 1e-24
Identities = 66/177 (37%), Positives = 105/177 (59%), Gaps = 7/177 (3%)
Frame = +1
Query: 290 IFVSAGFSES----IGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAG--TCLGCFLA 343
IF +AG ++ T+A+ K ++ ++L+DK GR+PLL++S G CL C A
Sbjct: 7 IFQAAGIEDNSKLLAATVAVGVAKTIFILVAIILIDKLGRKPLLMISTIGMTVCLFCMGA 186
Query: 344 ALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVT 403
LA L +G + LA++ V V +S+G+G + WV+ SEIFP+ V+ A +L
Sbjct: 187 TLALLGKG-----SFAIALAILFVCGNVAFFSVGLGPVCWVLTSEIFPLRVRAQASALGA 351
Query: 404 FFNWLCSWIVSYAF-NFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQ 459
N +CS +V+ +F + + S GTFF+F++I + FV LVPETKG++LE+I+
Sbjct: 352 VANRVCSGLVAMSFLSVSEAISVAGTFFVFAAISALAIAFVVTLVPETKGKSLEQIE 522
>TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(54%)
Length = 1142
Score = 108 bits (270), Expect = 5e-24
Identities = 76/262 (29%), Positives = 131/262 (49%), Gaps = 6/262 (2%)
Frame = +3
Query: 203 DSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYM 262
D+P L + G L+E + L+++RG + ++ E E+ + + + ++ + L + +
Sbjct: 3 DTPNSLIERGRLEEGKTVLKKIRGTD-NIELEFQELVEASR-VAKEVKHPFRNLLKRRNR 176
Query: 263 TSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMV---AVKIPVTVLGVLL 319
L I + L I QQF GIN I+FYA +F + GF + V AV + TV+ +
Sbjct: 177 PQLVISIALQIFQQFTGINAIMFYAPVLFNTLGFKNDASLYSAVITGAVNVLSTVVSIYS 356
Query: 320 MDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSP---ILALVGVLVYVGSYSL 376
+DK GRR LLL + L + A+ ++ +S IL +V V +V S++
Sbjct: 357 VDKLGRRMLLLEAGVQMFLSQVVIAIILGIKVTDHSDDLSKGIAILVVVMVCTFVSSFAW 536
Query: 377 GMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSIC 436
G + W+I SE FP+ + + S+ N L +++++ AF ++ G F FS
Sbjct: 537 SWGPLGWLIPSETFPLETRSAGQSVTVCVNLLFTFVIAQAFLSMLCHFKFGIFLFFSGWV 716
Query: 437 GFTVLFVAKLVPETKGRTLEEI 458
+FV L+PETK +EE+
Sbjct: 717 LVMSVFVLFLLPETKNVPIEEM 782
>TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%)
Length = 993
Score = 104 bits (259), Expect = 1e-22
Identities = 66/210 (31%), Positives = 111/210 (52%), Gaps = 3/210 (1%)
Frame = +3
Query: 256 LFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPVTVL 315
L +Y + IG L LQQ GIN + ++++++F S G I + + +V+
Sbjct: 123 LIYGRYFRVMFIGSTLFALQQLSGINAVFYFSSTVFESFGVPSDIANSCVGVCNLLGSVV 302
Query: 316 GVLLMDKSGRRPLLLLS--AAGTCLGCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGS 373
++LMDK GR+ LLL S G +G + A + G S L++ G+L++V S
Sbjct: 303 AMILMDKLGRKVLLLGSFLGMGLSMGLQVIAASSFASGFG-----SMYLSVGGMLLFVLS 467
Query: 374 YSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFF-IF 432
++ G G +P +IMSEI P N++ A ++ +W+ ++ V F L+ + IF
Sbjct: 468 FAFGAGPVPCLIMSEILPSNIRAKAMAICLAVHWVINFFVGLFFLRLLELIGAQLLYSIF 647
Query: 433 SSICGFTVLFVAKLVPETKGRTLEEIQASL 462
C V+FV K + ETKG++L+EI+ +L
Sbjct: 648 GFCCLIAVVFVKKNILETKGKSLQEIEIAL 737
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,815,932
Number of Sequences: 63676
Number of extensions: 325708
Number of successful extensions: 2693
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 2555
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2622
length of query: 471
length of database: 12,639,632
effective HSP length: 101
effective length of query: 370
effective length of database: 6,208,356
effective search space: 2297091720
effective search space used: 2297091720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0544.5


