

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
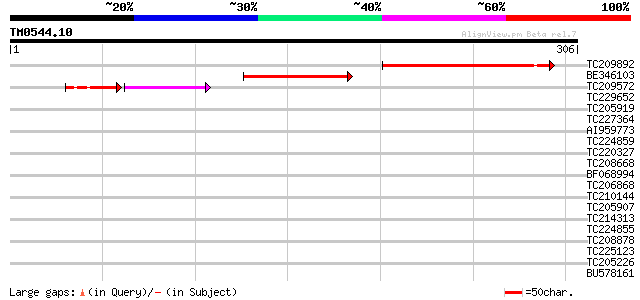
Query= TM0544.10
(306 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209892 UP|Q7PHQ2 (Q7PHQ2) ENSANGP00000022785 (Fragment), parti... 96 2e-20
BE346103 70 2e-12
TC209572 35 3e-06
TC229652 35 0.056
TC205919 similar to UP|Q94F39 (Q94F39) AT3g29575/MWE13_2, partia... 33 0.21
TC227364 similar to UP|Q9SYH5 (Q9SYH5) F15I1.27, partial (8%) 32 0.47
AI959773 homologue to GP|4467130|emb| glycosyltransferase like p... 32 0.47
TC224859 weakly similar to UP|Q7TP79 (Q7TP79) Aa2-245, partial (3%) 31 0.61
TC220327 similar to UP|Q8RY07 (Q8RY07) Serine/threonine protein ... 31 0.61
TC208668 homologue to UP|Q9NGW8 (Q9NGW8) Developmental protein D... 31 0.61
BF068994 similar to GP|15010768|gb| AT3g06130/F28L1_7 {Arabidops... 31 0.80
TC206868 similar to UP|PEX2_ARATH (Q9CA86) Peroxisome assembly p... 31 0.80
TC210144 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, part... 30 1.0
TC205907 similar to UP|Q6K6F2 (Q6K6F2) Hydroxyproline-rich glyco... 30 1.4
TC214313 weakly similar to UP|Q9SFZ3 (Q9SFZ3) T22C5.11, partial ... 30 1.4
TC224855 similar to UP|KHL1_HUMAN (Q9NR64) Kelch-like protein 1,... 30 1.4
TC208878 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper p... 30 1.4
TC225123 similar to UP|Q9FJA6 (Q9FJA6) 40S ribosomal protein S3 ... 30 1.8
TC205226 similar to GB|AAO11561.1|27363284|BT002645 At2g25520/F1... 29 2.3
BU578161 similar to GP|21702253|emb photosystem I subunit O {Ara... 29 2.3
>TC209892 UP|Q7PHQ2 (Q7PHQ2) ENSANGP00000022785 (Fragment), partial (14%)
Length = 582
Score = 96.3 bits (238), Expect = 2e-20
Identities = 43/93 (46%), Positives = 62/93 (66%)
Frame = +1
Query: 202 GLASRVFLMKWFKWTVEQFDLLNNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPD 261
GL RVFLM+W+KWTV+QF+ LNN GNG VM+ G GG YSL +HH + E+ +GL+ +
Sbjct: 1 GLTLRVFLMRWYKWTVQQFESLNNMGNGNMLVMEKGFGGRYSLLMHHGEEELRRFGLTDE 180
Query: 262 MIADQKWRAHATKGAWHDQGPLSLDTFFDHLHD 294
M+ DQ+W A + P+ +++FF HL +
Sbjct: 181 MLIDQEWHKFAKPADLNYDCPM-VNSFFPHLSE 276
>BE346103
Length = 184
Score = 69.7 bits (169), Expect = 2e-12
Identities = 31/59 (52%), Positives = 44/59 (74%)
Frame = +2
Query: 127 REQDFGNFQVQERINALKETRQRFGRFFFRFPDGESAADVFDRVSSFLESLWRDIDMNR 185
REQD+GNFQ ++ + K RQR+GRFF+R P GESA DV+DR++ +E+L DI++ R
Sbjct: 2 REQDWGNFQHRDEMRVEKALRQRYGRFFYRLPYGESAVDVYDRITGCIETLRTDINIGR 178
>TC209572
Length = 370
Score = 35.4 bits (80), Expect(2) = 3e-06
Identities = 23/47 (48%), Positives = 25/47 (52%), Gaps = 1/47 (2%)
Frame = -2
Query: 63 TPQGITQARIAGARIRHVISSTS-STDWRVYFYVSPYARTRSTLREI 108
T QG+ A R VI S S DWRV F VS YARTRS E+
Sbjct: 255 TAQGMA*ALRPDKHFRRVIGSDDYSPDWRV*F*VSTYARTRSMFYEL 115
Score = 32.7 bits (73), Expect(2) = 3e-06
Identities = 19/30 (63%), Positives = 22/30 (73%)
Frame = -3
Query: 31 LPKRIILVRHGESQGNLDPSAYTTTPDHKI 60
L KRI L+ HGE QGN D + YTTTPD+ I
Sbjct: 347 LEKRI-LM*HGE-QGN*DTTTYTTTPDYSI 264
>TC229652
Length = 1485
Score = 34.7 bits (78), Expect = 0.056
Identities = 12/25 (48%), Positives = 19/25 (76%)
Frame = +3
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNH 26
HNHNNND+D+NNN +++ N ++
Sbjct: 6 HNHNNNDDDNNNNEIIKEVVNSGSN 80
Score = 28.9 bits (63), Expect = 3.0
Identities = 9/15 (60%), Positives = 15/15 (100%)
Frame = +3
Query: 3 NHNNNDNDDNNNHHQ 17
NHN+N+NDD+NN+++
Sbjct: 3 NHNHNNNDDDNNNNE 47
>TC205919 similar to UP|Q94F39 (Q94F39) AT3g29575/MWE13_2, partial (51%)
Length = 1558
Score = 32.7 bits (73), Expect = 0.21
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +3
Query: 2 HNHNNNDNDDNNNHHQQQIP 21
+N+NNN+N +NNNHH +P
Sbjct: 393 NNNNNNNNSNNNNHHNLPMP 452
Score = 28.5 bits (62), Expect = 4.0
Identities = 9/16 (56%), Positives = 15/16 (93%)
Frame = +3
Query: 1 MHNHNNNDNDDNNNHH 16
++N+NNN+N+ NNN+H
Sbjct: 387 VNNNNNNNNNSNNNNH 434
>TC227364 similar to UP|Q9SYH5 (Q9SYH5) F15I1.27, partial (8%)
Length = 1376
Score = 31.6 bits (70), Expect = 0.47
Identities = 13/33 (39%), Positives = 23/33 (69%)
Frame = -3
Query: 1 MHNHNNNDNDDNNNHHQQQIPNQNNHKPSVLPK 33
+HN+NNN+N++NNN++ N ++ +P PK
Sbjct: 1308 LHNNNNNNNNNNNNNN-----NNSSSRPPPPPK 1225
>AI959773 homologue to GP|4467130|emb| glycosyltransferase like protein
{Arabidopsis thaliana}, partial (17%)
Length = 455
Score = 31.6 bits (70), Expect = 0.47
Identities = 22/63 (34%), Positives = 27/63 (41%), Gaps = 11/63 (17%)
Frame = +1
Query: 147 RQRFGRFFFRFPDGESAADVFD---RVSSFLE--------SLWRDIDMNRLNHDPSNDLN 195
+ F R D E A D FD +S LE S W D + HDPS+ LN
Sbjct: 43 KHNFSRVIILEDDMEIAPDFFDYFEAAASLLEKDKSIMAVSSWNDNGQKQFVHDPSSALN 222
Query: 196 LII 198
L+I
Sbjct: 223 LVI 231
>TC224859 weakly similar to UP|Q7TP79 (Q7TP79) Aa2-245, partial (3%)
Length = 843
Score = 31.2 bits (69), Expect = 0.61
Identities = 21/61 (34%), Positives = 30/61 (48%), Gaps = 5/61 (8%)
Frame = +2
Query: 5 NNNDNDDNNNHHQQQIPNQNNHKPSVLPKRIILVRHGESQGNLDPSAY-----TTTPDHK 59
NNN+ND+ NN++ Q P +KP L + LV+ +L+P Y TP H
Sbjct: 47 NNNNNDNKNNNNNNQYPQSALYKPK*LLE--ALVKKV*KASSLNPFFYLPQTIVITPVHL 220
Query: 60 I 60
I
Sbjct: 221 I 223
>TC220327 similar to UP|Q8RY07 (Q8RY07) Serine/threonine protein kinase pk23,
partial (20%)
Length = 780
Score = 31.2 bits (69), Expect = 0.61
Identities = 15/30 (50%), Positives = 20/30 (66%)
Frame = -1
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSVL 31
+N+NNNDN++NNN N NN K S+L
Sbjct: 516 YNNNNNDNNNNNN-------NNNNKKISLL 448
>TC208668 homologue to UP|Q9NGW8 (Q9NGW8) Developmental protein DG1037
(Fragment), partial (4%)
Length = 1021
Score = 31.2 bits (69), Expect = 0.61
Identities = 12/25 (48%), Positives = 18/25 (72%)
Frame = -2
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHK 27
N+NNN+N++NNN++ N NN K
Sbjct: 279 NNNNNNNNNNNNNNNNNNNNNNNIK 205
Score = 29.6 bits (65), Expect = 1.8
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Frame = -2
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKP--SVLPKRIILVR 39
+N+NNN+N++NNN++ N HK S L K+I ++
Sbjct: 273 NNNNNNNNNNNNNNNNNNNNNIKLHKKTGSKLNKKIAPIK 154
Score = 28.5 bits (62), Expect = 4.0
Identities = 10/23 (43%), Positives = 17/23 (73%)
Frame = -2
Query: 2 HNHNNNDNDDNNNHHQQQIPNQN 24
+N+NNN+N++NNN++ N N
Sbjct: 279 NNNNNNNNNNNNNNNNNNNNNNN 211
>BF068994 similar to GP|15010768|gb| AT3g06130/F28L1_7 {Arabidopsis
thaliana}, partial (4%)
Length = 240
Score = 30.8 bits (68), Expect = 0.80
Identities = 14/30 (46%), Positives = 18/30 (59%), Gaps = 2/30 (6%)
Frame = +1
Query: 2 HNHNNNDNDDNNNHHQQQIPNQ--NNHKPS 29
+N+NNN N +N NH Q+ N NNH S
Sbjct: 52 NNNNNNHNHNNQNHLADQLKNMQINNHPKS 141
>TC206868 similar to UP|PEX2_ARATH (Q9CA86) Peroxisome assembly protein 2
(Peroxin-2) (AthPEX2) (Pex2p), partial (86%)
Length = 1490
Score = 30.8 bits (68), Expect = 0.80
Identities = 9/26 (34%), Positives = 22/26 (84%)
Frame = -3
Query: 1 MHNHNNNDNDDNNNHHQQQIPNQNNH 26
++N+NNN+N++NNN++++++ + H
Sbjct: 1284 VNNNNNNNNNNNNNNNKEKVRGKEYH 1207
>TC210144 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, partial (54%)
Length = 1167
Score = 30.4 bits (67), Expect = 1.0
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +2
Query: 177 LWRDIDMNRLNHDPSNDLNLIIVSHGLASRVFLMKWFKWTV 217
+W D N +HDP +L L+I SH + +F M + +V
Sbjct: 932 MWN*CDWNATSHDPWENLFLLIRSHQILFYIFFMLFVSTSV 1054
>TC205907 similar to UP|Q6K6F2 (Q6K6F2) Hydroxyproline-rich
glycoprotein-like, partial (27%)
Length = 833
Score = 30.0 bits (66), Expect = 1.4
Identities = 17/46 (36%), Positives = 21/46 (44%)
Frame = -3
Query: 224 NNFGNGEFRVMQLGSGGEYSLAVHHTDREMHEWGLSPDMIADQKWR 269
N FGN R LG G + R HE G+ PD +D +WR
Sbjct: 222 NGFGNLRRRRCDLGGGNGVVVGGFGCRRRRHEDGVRPD-FSDHRWR 88
>TC214313 weakly similar to UP|Q9SFZ3 (Q9SFZ3) T22C5.11, partial (7%)
Length = 618
Score = 30.0 bits (66), Expect = 1.4
Identities = 10/19 (52%), Positives = 16/19 (83%)
Frame = -3
Query: 2 HNHNNNDNDDNNNHHQQQI 20
HN+NNN+N++NNN++ I
Sbjct: 67 HNNNNNNNNNNNNNNNNNI 11
Score = 27.7 bits (60), Expect = 6.8
Identities = 13/28 (46%), Positives = 20/28 (71%)
Frame = -3
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHKPSV 30
+HNNN+N++NNN++ N NN+ SV
Sbjct: 70 SHNNNNNNNNNNNN-----NNNNNI*SV 2
>TC224855 similar to UP|KHL1_HUMAN (Q9NR64) Kelch-like protein 1, partial
(3%)
Length = 885
Score = 30.0 bits (66), Expect = 1.4
Identities = 14/34 (41%), Positives = 22/34 (64%)
Frame = -2
Query: 3 NHNNNDNDDNNNHHQQQIPNQNNHKPSVLPKRII 36
N+NNNDN +NNN++ Q P +KP L + ++
Sbjct: 857 NNNNNDNKNNNNNN-NQYPQSALYKPK*LFEALV 759
>TC208878 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper protein,
partial (46%)
Length = 701
Score = 30.0 bits (66), Expect = 1.4
Identities = 13/24 (54%), Positives = 17/24 (70%)
Frame = +1
Query: 5 NNNDNDDNNNHHQQQIPNQNNHKP 28
N++DN++N NHHQQQ HKP
Sbjct: 55 NDDDNNNNLNHHQQQQYPFYVHKP 126
Score = 27.3 bits (59), Expect = 8.9
Identities = 13/31 (41%), Positives = 18/31 (57%), Gaps = 3/31 (9%)
Frame = +1
Query: 7 NDNDDNNN---HHQQQIPNQNNHKPSVLPKR 34
ND+D+NNN H QQQ P + P +P +
Sbjct: 55 NDDDNNNNLNHHQQQQYPFYVHKPPQSVPNQ 147
>TC225123 similar to UP|Q9FJA6 (Q9FJA6) 40S ribosomal protein S3
(AT5g35530/MOK9_14), partial (44%)
Length = 604
Score = 29.6 bits (65), Expect = 1.8
Identities = 13/28 (46%), Positives = 16/28 (56%)
Frame = -1
Query: 4 HNNNDNDDNNNHHQQQIPNQNNHKPSVL 31
HN+ +N NHH +Q PN N KP L
Sbjct: 157 HNDLLKPNNQNHHTKQHPNITNLKPKSL 74
>TC205226 similar to GB|AAO11561.1|27363284|BT002645 At2g25520/F13B15.18
{Arabidopsis thaliana;} , partial (52%)
Length = 970
Score = 29.3 bits (64), Expect = 2.3
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = -1
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHKPSV 30
+N NNN++DDNNN+ + + N K ++
Sbjct: 736 YNDNNNNDDDNNNYLIAEYITEQNQKQNL 650
>BU578161 similar to GP|21702253|emb photosystem I subunit O {Arabidopsis
thaliana}, partial (43%)
Length = 409
Score = 29.3 bits (64), Expect = 2.3
Identities = 10/26 (38%), Positives = 21/26 (80%)
Frame = -2
Query: 2 HNHNNNDNDDNNNHHQQQIPNQNNHK 27
+N+NNN+N++NNN++ ++ + N+K
Sbjct: 342 NNNNNNNNNNNNNNNNKKE*KKKNYK 265
Score = 28.1 bits (61), Expect = 5.2
Identities = 12/27 (44%), Positives = 19/27 (69%)
Frame = -2
Query: 1 MHNHNNNDNDDNNNHHQQQIPNQNNHK 27
++N+NNN+N++NNN N NN+K
Sbjct: 351 LNNNNNNNNNNNNN-------NNNNNK 292
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,730,275
Number of Sequences: 63676
Number of extensions: 211294
Number of successful extensions: 2197
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 1518
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1843
length of query: 306
length of database: 12,639,632
effective HSP length: 97
effective length of query: 209
effective length of database: 6,463,060
effective search space: 1350779540
effective search space used: 1350779540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0544.10


