

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
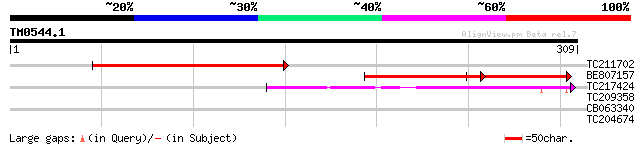
Query= TM0544.1
(309 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211702 192 2e-49
BE807157 114 6e-26
TC217424 52 3e-07
TC209358 similar to UP|P92987 (P92987) Myosin heavy chain-like p... 28 4.0
CB063340 similar to GP|15010610|gb| At1g05140/YUP8H12_25 {Arabid... 28 6.9
TC204674 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein... 27 9.0
>TC211702
Length = 465
Score = 192 bits (488), Expect = 2e-49
Identities = 96/107 (89%), Positives = 99/107 (91%)
Frame = +2
Query: 46 KELLKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAESLLSPVDS 105
KEL KGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDEL+LNGIE SLLSPVD+
Sbjct: 2 KELAKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELRLNGIETLSLLSPVDT 181
Query: 106 TLGSIERTLQIAAIAGGLTAWNAFGISPQQLFYISLGLLFLWTLDTV 152
TLGSIER LQIAAI GGL AWNAF ISPQQ+FYISLGLLFLWTLD V
Sbjct: 182 TLGSIERNLQIAAIVGGLAAWNAFAISPQQIFYISLGLLFLWTLDAV 322
>BE807157
Length = 344
Score = 114 bits (285), Expect = 6e-26
Identities = 56/66 (84%), Positives = 62/66 (93%)
Frame = +3
Query: 194 ILPKGYTLSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAGVCT 253
ILP+GYT+SSLDAL+K GSLN+QAG+AFVDFEF EEVN+GKVSA TLNKFSCIALAGV T
Sbjct: 3 ILPRGYTISSLDALQKVGSLNIQAGTAFVDFEFQEEVNSGKVSATTLNKFSCIALAGVST 182
Query: 254 EYLIYG 259
EYLIYG
Sbjct: 183 EYLIYG 200
Score = 89.0 bits (219), Expect = 3e-18
Identities = 45/57 (78%), Positives = 46/57 (79%)
Frame = +1
Query: 250 GVCTEYLIYGFSEGGLDDIRKLDSLLSGLGFTQKKVDSQVRWSVLNTVLLLRRHEAA 306
G L FSEGGLDDIRKLD LL GLGFTQKK DSQVRWS+LNTVLLLRRHEAA
Sbjct: 172 GCLLNILYMEFSEGGLDDIRKLDLLLKGLGFTQKKTDSQVRWSLLNTVLLLRRHEAA 342
>TC217424
Length = 1425
Score = 52.0 bits (123), Expect = 3e-07
Identities = 49/174 (28%), Positives = 78/174 (44%), Gaps = 6/174 (3%)
Frame = +1
Query: 141 LGLLFLWTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGHFLIAYLVGILPKGYT 200
+G + L L S GL + + F R+ +HEA HFLIAYL+G+ Y
Sbjct: 727 IGSISLVVLAVGSISPGLLQAAIGSFSTLFPDD-QERIARHEAAHFLIAYLLGLPIFDY- 900
Query: 201 LSSLDALKKDGSLNVQAGSAFVDFEFLEEVNAGKVSAKTLNKFSCIALAGVCTEYLIYGF 260
SLD K+ +L +D + + +G++ A+ L++ + +++AG+ E L Y
Sbjct: 901 --SLDIGKEHVNL--------IDERLEKLIYSGQLDAQELDRLAVVSMAGLAAEGLTYDK 1050
Query: 261 SEGGLDDIRKLDSLLSGLGFTQKKVDSQ--VRWSVLNTVLLLRR----HEAARA 308
G D+ L ++ K Q RW+VL LL+ HEA A
Sbjct: 1051VVGQSADLFSLQRFINRTKPPLSKDQQQNLTRWAVLFAASLLKNNKVTHEALMA 1212
>TC209358 similar to UP|P92987 (P92987) Myosin heavy chain-like protein,
partial (17%)
Length = 740
Score = 28.5 bits (62), Expect = 4.0
Identities = 26/91 (28%), Positives = 37/91 (40%)
Frame = +1
Query: 38 RQLLEKLDKELLKGDDRAALALVKDLQGKPDGLRCFGAARQVPQRLYTLDELKLNGIEAE 97
R+ +E+L ++LL+ D+ L+K + D + F A LDELK E E
Sbjct: 7 REQVEELQRKLLEKDE-----LLKSAENTRDQMNVFNAK---------LDELKHQASEKE 144
Query: 98 SLLSPVDSTLGSIERTLQIAAIAGGLTAWNA 128
SLL L + L A W A
Sbjct: 145 SLLKYTQQQLSDAKIKLADKQAALEKIQWEA 237
>CB063340 similar to GP|15010610|gb| At1g05140/YUP8H12_25 {Arabidopsis
thaliana}, partial (18%)
Length = 421
Score = 27.7 bits (60), Expect = 6.9
Identities = 14/39 (35%), Positives = 21/39 (52%)
Frame = +3
Query: 178 VIQHEAGHFLIAYLVGILPKGYTLSSLDALKKDGSLNVQ 216
++ HE+GHFL A L GI + + L K + NV+
Sbjct: 288 IVVHESGHFLAASLQGIHVSKFAVGFGPILAKFNAKNVE 404
>TC204674 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein, partial
(98%)
Length = 635
Score = 27.3 bits (59), Expect = 9.0
Identities = 13/39 (33%), Positives = 21/39 (53%)
Frame = +2
Query: 147 WTLDTVSYGGGLGNLVVDTIGHSFSKKYHNRVIQHEAGH 185
WT +TV++ G DT+ ++S HN V +AG+
Sbjct: 158 WTFNTVTWPQGKRFTAGDTLAFNYSPGAHNVVAVSKAGY 274
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,770,493
Number of Sequences: 63676
Number of extensions: 158608
Number of successful extensions: 714
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 712
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 714
length of query: 309
length of database: 12,639,632
effective HSP length: 97
effective length of query: 212
effective length of database: 6,463,060
effective search space: 1370168720
effective search space used: 1370168720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0544.1


