

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0541.9
(266 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
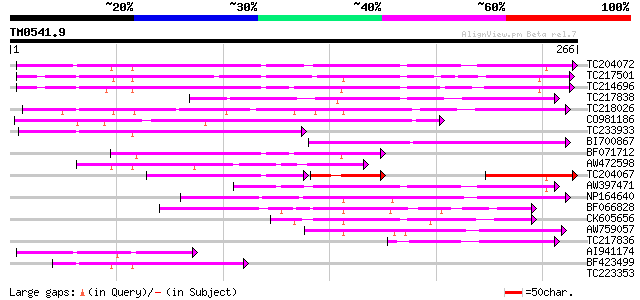
Score E
Sequences producing significant alignments: (bits) Value
TC204072 UP|LEC_SOYBN (P05046) Lectin precursor (Agglutinin) (SB... 167 6e-42
TC217501 similar to UP|Q703U4 (Q703U4) Lectin precursor, partial... 156 8e-39
TC214696 similar to UP|Q8L683 (Q8L683) Lectin precursor, partial... 152 2e-37
TC217838 weakly similar to PIR|A03357|CVJBP concanavalin A precu... 124 6e-29
TC218026 weakly similar to UP|Q9ZTA9 (Q9ZTA9) Mannose lectin, pa... 122 2e-28
CO981186 102 2e-22
TC233933 similar to UP|Q43563 (Q43563) Lectin precursor, partial... 94 5e-20
BI700867 similar to GP|1755066|gb|A lectin precursor {Sophora ja... 86 2e-17
BF071712 weakly similar to GP|22208832|em lectin {Vigna linearis... 83 1e-16
AW472598 similar to GP|3114258|pdb Soybean Agglutinin Complexed ... 81 4e-16
TC204067 lectin [Glycine max] 63 2e-15
AW397471 similar to GP|3114258|pdb Soybean Agglutinin Complexed ... 69 2e-12
NP164640 receptor-like protein kinase 69 2e-12
BF066828 weakly similar to PIR|A31972|A319 lectin DB58 precursor... 65 3e-11
CK605656 60 1e-09
AW759057 weakly similar to GP|6650223|gb| receptor-like protein ... 53 1e-07
TC217836 similar to UP|Q9M7M4 (Q9M7M4) Mannose lectin FRIL (Frag... 47 9e-06
AI941174 similar to SP|P05046|LEC_S Lectin precursor (Agglutinin... 44 1e-04
BF423499 weakly similar to GP|4115545|dbj| lectin {Robinia pseud... 43 2e-04
TC223353 weakly similar to UP|Q9SEW3 (Q9SEW3) Receptor-like prot... 39 0.002
>TC204072 UP|LEC_SOYBN (P05046) Lectin precursor (Agglutinin) (SBA), complete
Length = 1086
Score = 167 bits (422), Expect = 6e-42
Identities = 100/267 (37%), Positives = 155/267 (57%), Gaps = 4/267 (1%)
Frame = +1
Query: 4 MSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFA-SNGVLELTKT-AYGVP 61
++L ++L+ L A++ + +F++ F +P ++ +GDA S+G L+L K G P
Sbjct: 172 LTLTLVLVLLTSKANSAETVSFSWNKF-VPKQPNMILQGDAIVTSSGKLQLNKVDENGTP 348
Query: 62 LPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPK 121
PSS GRA ++ P+H+WDK+TG++A+F SF + DG+AFF+AP ++K
Sbjct: 349 KPSSLGRALYSTPIHIWDKETGSVASFAASFNFTFYAPDTKRLADGLAFFLAPIDTK--P 522
Query: 122 NSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTVPWRTG 181
+ G+LGLF+++ + D Q+VAVEFDT+ ++WDP HIGI++NSI S KT W
Sbjct: 523 QTHAGYLGLFNENESGD----QVVAVEFDTFRNSWDPPNPHIGINVNSIRSIKTTSWDLA 690
Query: 182 NSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVG 241
N+ A ++Y+ T L + Q+ + LS VVDL+T LPEWVR+G
Sbjct: 691 NN---KVAKVLITYDASTSLLVASLVYPSQR-------TSNILSDVVDLKTSLPEWVRIG 840
Query: 242 FSGATGQLV--EQHRIHVWDFKSSFTY 266
FS ATG + E H + W F S+ +
Sbjct: 841 FSAATGLDIPGESHDVLSWSFASNLPH 921
>TC217501 similar to UP|Q703U4 (Q703U4) Lectin precursor, partial (79%)
Length = 1011
Score = 156 bits (395), Expect = 8e-39
Identities = 111/268 (41%), Positives = 149/268 (55%), Gaps = 6/268 (2%)
Frame = +2
Query: 4 MSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFAS-NGVLELTKT-AYGVP 61
++ F++L++ A++ N+ +FT F P +++ F+GDA S +GVL LTK + VP
Sbjct: 59 LAFFLVLLTK---ANSTNTVSFTVSKFS-PRQQNLIFQGDAAISPSGVLRLTKVDSIDVP 226
Query: 62 LPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPK 121
S GRA +A P+ +WD +TG +A++ TSF V S DG+AFF+AP SK
Sbjct: 227 TTGSLGRALYATPIQIWDSETGKVASWATSFKFKV--FSPNKTADGLAFFLAPVGSK--P 394
Query: 122 NSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADT-WDPFTSHIGIDINSIISSKTVPWRT 180
S GGFLGLF+ DS S Q VAVEFDTY + WDP HIGID+NSI S KT W
Sbjct: 395 QSKGGFLGLFNSDSKNKSVQT--VAVEFDTYYNAKWDPANRHIGIDVNSIKSVKTASWGL 568
Query: 181 GNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRV 240
N A ++Y+ T S+LV + K S I LS V L++ LPEWV +
Sbjct: 569 ANG---QIAQILITYDADT---SLLVASL-IHPSRKTSYI---LSETVSLKSNLPEWVNI 718
Query: 241 GFSGATG---QLVEQHRIHVWDFKSSFT 265
GFS TG VE H + W F S +
Sbjct: 719 GFSATTGLNKGFVETHDVFSWSFASKLS 802
>TC214696 similar to UP|Q8L683 (Q8L683) Lectin precursor, partial (80%)
Length = 1701
Score = 152 bits (383), Expect = 2e-37
Identities = 103/268 (38%), Positives = 146/268 (54%), Gaps = 6/268 (2%)
Frame = +3
Query: 4 MSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDA-FASNGVLELTKT-AYGVP 61
++ F++L++ AH+ ++ +FTF F P +I + DA +S+GVL+LTK + GVP
Sbjct: 78 LAFFLVLLTK---AHSTDTVSFTFNKFN-PVQPNIMLQKDASISSSGVLQLTKVGSNGVP 245
Query: 62 LPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPK 121
S GRA +A P+ +WD +TG +A++ TSF ++ + DG+AFF+AP S+
Sbjct: 246 TSGSLGRALYAAPIQIWDSETGKVASWATSFKFNIFAPNKSNSADGLAFFLAPVGSQ--P 419
Query: 122 NSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYAD-TWDPFTSHIGIDINSIISSKTVPWRT 180
S GFLGLF +S Q VA+EFDT+++ WDP HIGID+NSI S KT W
Sbjct: 420 QSDDGFLGLF--NSPLKDKSLQTVAIEFDTFSNKKWDPANRHIGIDVNSIKSVKTASWGL 593
Query: 181 GNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRV 240
N A V+Y T L + I K + LS V+L++ LPEWV V
Sbjct: 594 SNG---QVAEILVTYNAATSLL--VASLIHPSKKTSY-----ILSDTVNLKSNLPEWVSV 743
Query: 241 GFSGATG---QLVEQHRIHVWDFKSSFT 265
GFS TG VE H + W F S +
Sbjct: 744 GFSATTGLHEGSVETHDVISWSFASKLS 827
>TC217838 weakly similar to PIR|A03357|CVJBP concanavalin A precursor - jack
bean {Canavalia ensiformis;} , partial (52%)
Length = 1202
Score = 124 bits (310), Expect = 6e-29
Identities = 76/176 (43%), Positives = 97/176 (54%), Gaps = 2/176 (1%)
Frame = +3
Query: 85 LAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQI 144
+A+F T FT + S+ GDG+AFFIAPF++KIP NS G LGLF D+ +
Sbjct: 18 VASFETDFTFSISSDSTT-PGDGLAFFIAPFDTKIPPNSGGSNLGLFPSDN--------V 170
Query: 145 VAVEFDTY--ADTWDPFTSHIGIDINSIISSKTVPWRTGNSLSTSAAFATVSYEPVTKTL 202
VAVEFDTY D DP HIGID+NSI+S T W N A +SY +K L
Sbjct: 171 VAVEFDTYPNRDKGDPDYRHIGIDVNSIVSKATARWEWQNG---KIATVHISYNSASKRL 341
Query: 203 SVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVW 258
+V T T +LS ++L +LPEWVRVG S +TGQ + + IH W
Sbjct: 342 TV--------AAFYPGTQTVTLSHDIELNKVLPEWVRVGLSASTGQQKQTNTIHSW 485
>TC218026 weakly similar to UP|Q9ZTA9 (Q9ZTA9) Mannose lectin, partial (72%)
Length = 1102
Score = 122 bits (306), Expect = 2e-28
Identities = 100/288 (34%), Positives = 138/288 (47%), Gaps = 31/288 (10%)
Frame = +3
Query: 7 FVILISLFMVAHNVNSE---TFTFPGFQLPNTKDITFEGDAFAS------NGVLELTKTA 57
F + M+ + VNS +F+F F + +D+ +GDA VL+LTK
Sbjct: 81 FTMATMFLMLLNRVNSADSLSFSFNNFS-EDQEDLILQGDATTGASSENDKNVLQLTKLD 257
Query: 58 -YGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTS-SGFHGDGIAFFIAPF 115
G P S GR + PVHLW K + ++ F T+FT + S DG+AFFIA
Sbjct: 258 DSGKPEFGSVGRVLYFAPVHLW-KSSQLVSTFETTFTFKISSASPDSVPADGLAFFIA-- 428
Query: 116 NSKIPKNSTGGFLGLFS-----KDSAFDSYQN-------------QIVAVEFDTYADT-- 155
+ + G LGLF K+S+ ++ +VAVEFDT+ +T
Sbjct: 429 SPDTTPGAGGQDLGLFPHLTSLKNSSSSHHRKVTRITGVKDLASEPVVAVEFDTFINTDI 608
Query: 156 WDPFTSHIGIDINSIISSKTVPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGL 215
DP HIGIDINSI S T W N + +A +SY +K L+V+ D
Sbjct: 609 GDPEYQHIGIDINSITSVTTTKWDWQNGKTVTAQ---ISYNSASKRLTVVASYPD----- 764
Query: 216 KFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDFKSS 263
+ SL + +DL TILPEWVRVGFS +TG E + + W F SS
Sbjct: 765 ---STPVSLYYDIDLFTILPEWVRVGFSASTGGAAEANTLLSWSFSSS 899
>CO981186
Length = 801
Score = 102 bits (254), Expect = 2e-22
Identities = 67/206 (32%), Positives = 106/206 (50%), Gaps = 4/206 (1%)
Frame = +1
Query: 3 LMSLFVILISLFMVAHNVNSETFTFPGF-QLPNTKDITFEGD-AFASNGVLELTKTAYGV 60
++++F++++++ S F F + K++ + GD A NG +EL Y
Sbjct: 115 VITVFLLVLAIPSPLKTAESLNFNITNFANSESAKNMLYVGDGAVNKNGSIELNIVDYDF 294
Query: 61 PLPSSSGRASFAEPVHLWDKKTGALAAFTT--SFTLDVQPTSSGFHGDGIAFFIAPFNSK 118
+ GRA + +P+ LWD +G + F+T +FT+D S + DG AF+IAP +
Sbjct: 295 RV----GRALYGQPLRLWDSSSGVVTDFSTRFTFTIDRGNNKSASYADGFAFYIAPHGYQ 462
Query: 119 IPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTVPW 178
IP N+ GG LF+ S +N ++AVEFDT+ T DP H+GID NS+ S T +
Sbjct: 463 IPPNAAGGTFALFNVTSNPFIPRNHVLAVEFDTFNGTIDPPFQHVGIDDNSLKSVATAKF 642
Query: 179 RTGNSLSTSAAFATVSYEPVTKTLSV 204
+L A V+Y +TL V
Sbjct: 643 DIDKNLXXKCN-ALVNYNASNRTLFV 717
>TC233933 similar to UP|Q43563 (Q43563) Lectin precursor, partial (7%)
Length = 422
Score = 94.4 bits (233), Expect = 5e-20
Identities = 52/136 (38%), Positives = 79/136 (57%), Gaps = 1/136 (0%)
Frame = +3
Query: 5 SLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKT-AYGVPLP 63
S+F++ L + + +S +F P F+ P+ +I +G A + GVL+LTK G P
Sbjct: 15 SVFLMTFLLLITSAKSDSFSFNLPRFE-PDALNILLDGSAKTTGGVLQLTKKDKRGNPTQ 191
Query: 64 SSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNS 123
S G ++F +HL D KTG +A F T F+ V + HGDG F++A + P NS
Sbjct: 192 HSVGLSAFYAALHLSDAKTGRVANFATEFSFVVNTKGAPLHGDGFTFYLASLDFDFPDNS 371
Query: 124 TGGFLGLFSKDSAFDS 139
+GGFLGLF+K +AF++
Sbjct: 372 SGGFLGLFNKKTAFNT 419
>BI700867 similar to GP|1755066|gb|A lectin precursor {Sophora japonica},
partial (19%)
Length = 430
Score = 85.9 bits (211), Expect = 2e-17
Identities = 44/123 (35%), Positives = 72/123 (57%)
Frame = +3
Query: 141 QNQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTVPWRTGNSLSTSAAFATVSYEPVTK 200
+N +VAVEFDT+ + DP T H+G+D NS+ S+ + ++L + ++Y T+
Sbjct: 12 ENHVVAVEFDTFIGSTDPPTKHVGVDDNSLTSAAFGNFDIDDNLGKKC-YTLITYTASTQ 188
Query: 201 TLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDF 260
TL V + + ++S S+ +DL+ ILPEWV +GFS +TG E++ I+ W+F
Sbjct: 189 TLFVSWSFKAKPASTNHNDNSSSFSYQIDLKKILPEWVNIGFSASTGLSTERNTIYSWEF 368
Query: 261 KSS 263
SS
Sbjct: 369 SSS 377
>BF071712 weakly similar to GP|22208832|em lectin {Vigna linearis}, partial
(43%)
Length = 421
Score = 82.8 bits (203), Expect = 1e-16
Identities = 52/131 (39%), Positives = 72/131 (54%), Gaps = 2/131 (1%)
Frame = +2
Query: 48 NGVLELTKTAY-GVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGD 106
+GVL++TK GVP S GRA F P+ +WD +TG +A + T F ++ D
Sbjct: 2 SGVLQITKVCISGVPTSGSFGRALFPAPIPIWDHETGNVAHWGTFFKFNIFSPHKSDSAD 181
Query: 107 GIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYAD-TWDPFTSHIGI 165
G+AFF AP SK S GFLGLF +SA VA+EFDT+++ WDP IG
Sbjct: 182 GLAFFFAPGGSK--PQSDHGFLGLF--NSALKDKSLPTVAIEFDTFSNKKWDPAHRLIGN 349
Query: 166 DINSIISSKTV 176
+N I ++ +
Sbjct: 350 GVNIINTASII 382
>AW472598 similar to GP|3114258|pdb Soybean Agglutinin Complexed With 2
4-Pentasaccharide, partial (49%)
Length = 421
Score = 81.3 bits (199), Expect = 4e-16
Identities = 52/141 (36%), Positives = 85/141 (59%), Gaps = 4/141 (2%)
Frame = +2
Query: 32 LPNTKDITFEGDAFA-SNGVLELTKT-AYGVPLPSSSGRASFAEPVHLWDKKTGAL--AA 87
LP + +GDA S+G L+L+K G P P S GRA ++ P+H+WDK+ ++ +A
Sbjct: 23 LPKQASMILQGDAIVTSSGKLQLSKVDENGTPKPLSLGRALYSTPIHIWDKEFVSVTASA 202
Query: 88 FTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAV 147
F + TL V T +G +AFF+AP ++K +++ +GL+++ Y +Q+VA
Sbjct: 203 FPSC*TLLVPDTRRLAYG--LAFFLAPIDTKPQTHASN--VGLYNEYE----YGDQVVAD 358
Query: 148 EFDTYADTWDPFTSHIGIDIN 168
E DT ++WDP +HIGI++N
Sbjct: 359 EIDTIRNSWDPPDTHIGINVN 421
>TC204067 lectin [Glycine max]
Length = 720
Score = 62.8 bits (151), Expect(2) = 2e-15
Identities = 28/76 (36%), Positives = 46/76 (59%)
Frame = +1
Query: 65 SSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNST 124
S GRA ++ P+H+WD +TG++A+F SF V + DG+AFF+AP +++ +
Sbjct: 13 SLGRALYSTPIHIWDSETGSVASFAASFNFTVYASDIANLADGLAFFLAPIDTQ--PQTR 186
Query: 125 GGFLGLFSKDSAFDSY 140
GG+LGL++ Y
Sbjct: 187 GGYLGLYNSTDTQQHY 234
Score = 49.3 bits (116), Expect = 2e-06
Identities = 23/45 (51%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Frame = +3
Query: 224 LSFVVDLRTILPEWVRVGFSGATGQLV--EQHRIHVWDFKSSFTY 266
LS V+DL+ LPEWVR+GFS TG V E H +H W F S+ +
Sbjct: 426 LSDVLDLKVALPEWVRIGFSATTGLNVASETHDVHSWSFSSNLPF 560
Score = 36.2 bits (82), Expect(2) = 2e-15
Identities = 19/35 (54%), Positives = 23/35 (65%)
Frame = +2
Query: 142 NQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTV 176
N I++VEFDT WD HIGI++NSI S K V
Sbjct: 224 NNIISVEFDT----WDSPNLHIGINVNSIRSIKLV 316
>AW397471 similar to GP|3114258|pdb Soybean Agglutinin Complexed With 2
4-Pentasaccharide, partial (71%)
Length = 562
Score = 68.9 bits (167), Expect = 2e-12
Identities = 51/155 (32%), Positives = 76/155 (48%), Gaps = 2/155 (1%)
Frame = +2
Query: 106 DGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTWDPFTSHIGI 165
DG+A F+AP ++K ++ LGLF+++ + D Q+ VE DT + WDP HIGI
Sbjct: 65 DGLACFLAPNDTKPQTHARD--LGLFNENESGD----QVDPVEIDTSRNCWDPPNPHIGI 226
Query: 166 DINSIISSKTVPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLS 225
+ N IIS KT W N+ + ++Y+ T L + Q+ + +L
Sbjct: 227 NGNCIISIKTTSWDLANN---KVSKVLITYDASTSLLDASLVYP*QR-------TSNTLF 376
Query: 226 FVVDLRTILPEWVRVGFSGATGQLV--EQHRIHVW 258
VD T EWV +GFS A G + E H + W
Sbjct: 377 DAVDQTTSRAEWVMIGFSAADGLDIPEESHDVLSW 481
>NP164640 receptor-like protein kinase
Length = 828
Score = 68.9 bits (167), Expect = 2e-12
Identities = 56/193 (29%), Positives = 89/193 (46%), Gaps = 10/193 (5%)
Frame = +1
Query: 81 KTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSY 140
K + +F+T+F + P G G AF I+ S + +LGL + + ++
Sbjct: 112 KNAKVVSFSTAFAFAIIPQYPKLGGHGFAFTISRSTS-LKDAYPSQYLGLLNPNDV-GNF 285
Query: 141 QNQIVAVEFDTYADT--WDPFTSHIGIDINSIISSKTVPW--------RTGNSLSTSAAF 190
N + AVEFDT D D +H+GI++N++ S+K+V + N S
Sbjct: 286 SNHLFAVEFDTVQDFEFGDINDNHVGINLNNMASNKSVEAAFFSRNNKQNLNLKSGEVTQ 465
Query: 191 ATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLV 250
A V Y+ + L V + + + LS+ VDL IL + + VGFS +TG L
Sbjct: 466 AWVDYDXLKNNLEVRLSTTSSKP------TSPILSYKVDLSPILQDSMYVGFSSSTGLLA 627
Query: 251 EQHRIHVWDFKSS 263
H I W FK++
Sbjct: 628 SSHYILGWSFKTN 666
>BF066828 weakly similar to PIR|A31972|A319 lectin DB58 precursor - horse
gram, partial (26%)
Length = 556
Score = 65.1 bits (157), Expect = 3e-11
Identities = 58/182 (31%), Positives = 87/182 (46%), Gaps = 5/182 (2%)
Frame = +2
Query: 71 FAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTGG--FL 128
+ P +W + G ++++ T F ++ + DG+AF AP + NS L
Sbjct: 8 YTVPFAIWYTQNGGVSSWATFFKFNIITPDNSNSSDGLAFICAPIGA----NSQFD**CL 175
Query: 129 GLFSKDSAFDSYQNQIVAVEFDTYADT-WDPFTSHIGIDINSIISSKTVP-WRTGNSLST 186
G+ ++ + D Y Q VA+EFDT+++ WDP HIGID+NSI S KT W
Sbjct: 176 GI*NR-TLKDRYL-QTVAIEFDTFSNKKWDPANRHIGIDVNSIKSVKTASRWLCNGQQED 349
Query: 187 SAAFATVSYEPVTKT-LSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGA 245
S V+Y T + L++ K S I + S +L + LPEW+ V FS
Sbjct: 350 S----FVTYNAATSL*VGNLIR-----PSKKTSFILSGHS---ELDSNLPEWLSVAFSST 493
Query: 246 TG 247
TG
Sbjct: 494 TG 499
>CK605656
Length = 628
Score = 59.7 bits (143), Expect = 1e-09
Identities = 46/130 (35%), Positives = 59/130 (45%), Gaps = 5/130 (3%)
Frame = -2
Query: 123 STGGFLGLFS---KDSAFDSYQNQIVAVEFDTYADT-WDPFTSHIGIDINSIISSKTVPW 178
S GF GLF+ KD F Q V +EFDT+++ WDP HIGI++NSI S KT W
Sbjct: 597 SDDGFFGLFNSPLKDKFF-----QTVGIEFDTFSNKKWDPANRHIGIEVNSIKSVKTASW 433
Query: 179 RTGNSLSTSAAFATVSYE-PVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEW 237
N VSY P + L+ + + F V+L+ PEW
Sbjct: 432 GLSNG---QVGGIFVSYNAPPGLLVGFLIPPSKKTSYILFDP--------VNLKGNFPEW 286
Query: 238 VRVGFSGATG 247
V VGF G
Sbjct: 285 VSVGFFAPPG 256
>AW759057 weakly similar to GP|6650223|gb| receptor-like protein kinase
{Glycine max}, partial (44%)
Length = 397
Score = 53.1 bits (126), Expect = 1e-07
Identities = 44/127 (34%), Positives = 61/127 (47%), Gaps = 4/127 (3%)
Frame = +3
Query: 139 SYQNQIVAVEFDTYADT--WDPFTSHIGIDINSIISSKTVPWR-TGNSL-STSAAFATVS 194
++ N I AVEFDT D D +H+GIDINS+ S+ + G +L S A V
Sbjct: 3 NFSNHIFAVEFDTVQDFEFGDINDNHVGIDINSMQSNTSANVSLVGLTLKSGKPILAWVD 182
Query: 195 YEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFSGATGQLVEQHR 254
Y+ +SV + + T L+F VDL + + + VGFS +TG L H
Sbjct: 183 YDSRLNLISVALSPNSSKPK------TPLLTFNVDLSPVFHDTMYVGFSASTGLLASFHY 344
Query: 255 IHVWDFK 261
I W FK
Sbjct: 345 I*GWSFK 365
>TC217836 similar to UP|Q9M7M4 (Q9M7M4) Mannose lectin FRIL (Fragment),
partial (25%)
Length = 437
Score = 47.0 bits (110), Expect = 9e-06
Identities = 28/81 (34%), Positives = 39/81 (47%)
Frame = +3
Query: 178 WRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEW 237
W+ G A +SY +K L+V Q T +LS ++L +LPEW
Sbjct: 63 WQNGK-----IATVHISYNSASKRLTVAAFYPGTQ--------TVTLSHDIELNKVLPEW 203
Query: 238 VRVGFSGATGQLVEQHRIHVW 258
VRVG S +TGQ + + IH W
Sbjct: 204 VRVGLSASTGQQKQTNTIHSW 266
>AI941174 similar to SP|P05046|LEC_S Lectin precursor (Agglutinin) (SBA).
[Soybean] {Glycine max}, partial (32%)
Length = 289
Score = 43.5 bits (101), Expect = 1e-04
Identities = 28/89 (31%), Positives = 50/89 (55%), Gaps = 4/89 (4%)
Frame = +2
Query: 4 MSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNG----VLELTKTAYG 59
++L + L+ L A++ + TF++ F LP ++ +GDA + + LTKTA
Sbjct: 29 ITLTMALVLLTSKANSA*TVTFSWNKF-LPKQPNMILQGDAMVTYSGKFNSIRLTKTA-- 199
Query: 60 VPLPSSSGRASFAEPVHLWDKKTGALAAF 88
P GRA ++ P+H+WDK+T + A++
Sbjct: 200 PQNPRVIGRALYSTPIHIWDKETDSAASY 286
>BF423499 weakly similar to GP|4115545|dbj| lectin {Robinia pseudoacacia},
partial (30%)
Length = 283
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/94 (29%), Positives = 48/94 (50%), Gaps = 2/94 (2%)
Frame = +2
Query: 21 NSETFTFPGFQLPNTKDITFEGDAFA-SNGVLELTKT-AYGVPLPSSSGRASFAEPVHLW 78
++++FTF F P +I + DA VL+LTK + VP S G A +A + W
Sbjct: 5 DTDSFTFNKFN-PVQPNIMLQKDAII*YYEVLQLTKVGSNSVPTSGSLGLALYAAQIQSW 181
Query: 79 DKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFI 112
D ++G + ++ T F ++ + DG+AF +
Sbjct: 182 DNESGKVNSWATFFKFNIYAPNQSNSDDGLAFIL 283
>TC223353 weakly similar to UP|Q9SEW3 (Q9SEW3) Receptor-like protein kinase
(Fragment), partial (17%)
Length = 714
Score = 39.3 bits (90), Expect = 0.002
Identities = 60/222 (27%), Positives = 81/222 (36%), Gaps = 9/222 (4%)
Frame = -1
Query: 1 MGLMSLFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGV 60
M SLF IL SL ++ +S T P + I GDA +
Sbjct: 651 MVAFSLF-ILCSLILLVQPSSSSTQQPPNL---DPDSIYLLGDAHVVTAADADSHVRLTR 484
Query: 61 PLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIP 120
P PSSSG EP+ D + +T F+ V HG G+ +A
Sbjct: 483 PAPSSSGILRRREPLAFADP-----TSLSTEFSFSVTG-----HGHGLLLVLA------- 355
Query: 121 KNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYAD--TWDPFTSHIGIDINSIISSKT--- 175
GG L + V VEFDT D DP +H+GID+ S +S
Sbjct: 354 ---AGGNLSNY-------------VGVEFDTSKDDNVGDPNANHVGIDVGSHVSVAVANV 223
Query: 176 ----VPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQK 213
+ G L+ A V YE +K L V + + QK
Sbjct: 222 SDVHLVLNNGEKLN-----AWVDYEASSKVLEVRLSKWGAQK 112
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,308,020
Number of Sequences: 63676
Number of extensions: 123323
Number of successful extensions: 730
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 699
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 700
length of query: 266
length of database: 12,639,632
effective HSP length: 96
effective length of query: 170
effective length of database: 6,526,736
effective search space: 1109545120
effective search space used: 1109545120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0541.9


