

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0490.11
(418 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
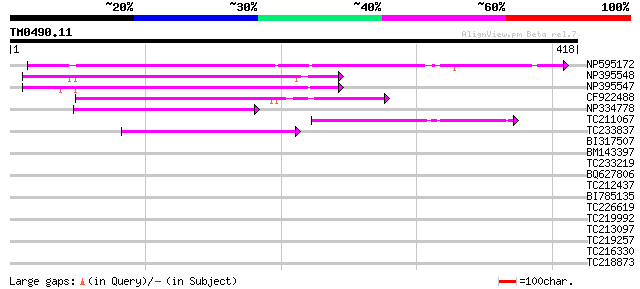
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 185 3e-47
NP395548 reverse transcriptase [Glycine max] 97 1e-20
NP395547 reverse transcriptase [Glycine max] 93 2e-19
CF922488 90 2e-18
NP334778 reverse transcriptase [Glycine max] 80 2e-15
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 55 4e-08
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 55 4e-08
BI317507 39 0.006
BM143397 32 0.53
TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance... 31 0.91
BQ627806 31 0.91
TC212437 weakly similar to UP|Q9SH20 (Q9SH20) F28K19.1, partial ... 30 2.0
BI785135 30 2.6
TC226619 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxid... 29 4.5
TC219992 similar to UP|Q75M40 (Q75M40) Unknow protein, partial (... 28 5.9
TC213097 28 5.9
TC219257 weakly similar to UP|Q6PLP6 (Q6PLP6) Cell wall protein ... 28 5.9
TC216330 similar to UP|Q6PMT1 (Q6PMT1) Methylthioribose kinase ... 28 7.7
TC218873 similar to UP|PSK3_ARATH (Q9M2Y0) Phytosulfokines 3 pre... 28 7.7
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 185 bits (470), Expect = 3e-47
Identities = 130/406 (32%), Positives = 197/406 (48%), Gaps = 7/406 (1%)
Frame = +1
Query: 14 QQLIKQGLIEPTKSEWACTAFYVNKRSEQVRQKKRLVVDYRPLNQFIKDDKFPLPKIYSL 73
Q+++ QG+I+P+ S ++ V K+ R DYR LN D FP+P + L
Sbjct: 1852 QEMLVQGIIQPSNSPFSLPILLVKKKDGSWR----FCTDYRALNAITVKDSFPMPTVDEL 2019
Query: 74 YSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKA 133
+ A FSK D++SG+ Q+ + P+DR KTAF + HY+W V+PFGL AP+ FQ
Sbjct: 2020 LDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCL 2199
Query: 134 MTKIFEPILHT-SLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEV 192
M KIF+ L LV+ DD+L++S + H K L+ +H + K EV
Sbjct: 2200 MNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEV 2379
Query: 193 DFLGMHFTEGKFVPQPHI-VEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKL 251
D+LG H G V + V+ + +P +KQ++ FLG+ Y R FI + L
Sbjct: 2380 DYLG-HKVSGLGVSMENTKVQAVLDWPTPN-NVKQLRGFLGLTGYYRRFIKSYANIAGPL 2553
Query: 252 SKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIPGDGKR-ILQTDASDHYWAAVLIEEI 310
+ +L+K++ W E A LKKA P L +P + IL+TDAS AVL
Sbjct: 2554 TDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVL---- 2721
Query: 311 DGQKLYCGHASGQFKD---AELHYHTTF-KETLAVKYGIKRFDFHLRGHHFEVQIDNSSF 366
GQ GH F + + + +E LA+ + +F +L G+ F ++ D S
Sbjct: 2722 -GQN---GHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSL 2889
Query: 367 PKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDYLSR 412
+++ +TP L F YDF +++ G N D LSR
Sbjct: 2890 KSLMDQSLQTPEQQAWLHK---FLGYDFKIEYKPGKDNQAADALSR 3018
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 97.4 bits (241), Expect = 1e-20
Identities = 78/256 (30%), Positives = 121/256 (46%), Gaps = 19/256 (7%)
Frame = +1
Query: 10 QEECQQLIKQGLIEP-TKSEWACTAFYVNKRSEQ--VRQKK------------RLVVDYR 54
++E +L++ GLI P + S W V+K+ +R +K +L +DYR
Sbjct: 4 RKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDYR 183
Query: 55 PLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHY 114
LN+ + D FPLP + + + + D G+ Q+ +DPKD+ K AF P +
Sbjct: 184 KLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGVF 363
Query: 115 QWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKKFLQQFYDIC 173
+ +PFGL AP+ FQ M IF I+ S+ V++DD +F ES K L+ C
Sbjct: 364 AYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQRC 543
Query: 174 SKHGVMLSSRKSQIATKEVDFLGMHF-TEGKFVPQPHI--VEELPKFPDEGLTIKQIQQF 230
+ ++L+ K +E LG T G V Q I +E+LP +K I+ F
Sbjct: 544 VETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLP----PPSNVKGIRSF 711
Query: 231 LGILNYIRDFIPHVSK 246
LG + R FI +K
Sbjct: 712 LGQARFYRRFIKDFTK 759
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 93.2 bits (230), Expect = 2e-19
Identities = 67/253 (26%), Positives = 120/253 (46%), Gaps = 16/253 (6%)
Frame = +1
Query: 10 QEECQQLIKQGLIEP-TKSEWACTAFYV----------NKRSEQVRQKK----RLVVDYR 54
++E +L++ GLI P + S W V N R+E + ++ R+ +DYR
Sbjct: 4 RKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDYR 183
Query: 55 PLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHY 114
LN+ + D +PLP + + + + D SG+ Q+ +DP+D+ KTAF P + +
Sbjct: 184 KLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSVF 363
Query: 115 QWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKKFLQQFYDIC 173
+ +PFGL A + FQ+ M IF+ ++ + V++DD F + L++ C
Sbjct: 364 AYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQRC 543
Query: 174 SKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHIVEELPKFPDEGLTIKQIQQFLGI 233
K ++L+ K +E LG ++ ++ + K P + +K I FLG
Sbjct: 544 EKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPP-VNVKGIHSFLGH 720
Query: 234 LNYIRDFIPHVSK 246
+ + R FI +K
Sbjct: 721 VGFYRRFIKDFTK 759
>CF922488
Length = 741
Score = 90.1 bits (222), Expect = 2e-18
Identities = 75/241 (31%), Positives = 114/241 (47%), Gaps = 9/241 (3%)
Frame = +3
Query: 49 LVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAFC 108
+ VDYR LN DKFPLP I L + FS D SG+ Q+ + P+D KT F
Sbjct: 30 MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDMEKTTFI 209
Query: 109 IPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKKFLQ 167
+ + + FGLK + +Q+AM +F ++H + VY+DD+++ S EE H L+
Sbjct: 210 TLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEEHLVNLR 389
Query: 168 QFYDICSKHGVMLSSRKSQIATKE---VDFL----GMHFTEGKFVPQPHIVEELPKFPDE 220
+ + K+ + L+ K K +DF+ G+ K ++ E+ K E
Sbjct: 390 KLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKV----KVILEMAKPHTE 557
Query: 221 GLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNA-PPWGKEQTEAIKFLKKAAQN 279
KQ+Q FLG LNYI FI + L +L KN W + A + +K+ N
Sbjct: 558 ----KQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLIN 725
Query: 280 P 280
P
Sbjct: 726 P 728
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 79.7 bits (195), Expect = 2e-15
Identities = 46/138 (33%), Positives = 75/138 (54%), Gaps = 1/138 (0%)
Frame = +3
Query: 48 RLVVDYRPLNQFIKDDKFPLPKIYSLYSYIKNAHIFSKFDMKSGFWQLGLDPKDRPKTAF 107
R+ VDYR LN+ D FPLP I L + + + +FS D SG+ Q+ + P+D KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 108 CIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPILHTSL-VYIDDVLLFSHDEESHKKFL 166
+ + V+ FGLK + + +AM +F+ ++H + Y+D+++ S EE H L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 167 QQFYDICSKHGVMLSSRK 184
Q + K+ + L+ RK
Sbjct: 363 QNLFGQLRKYRLRLNPRK 416
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 55.5 bits (132), Expect = 4e-08
Identities = 45/155 (29%), Positives = 78/155 (50%), Gaps = 2/155 (1%)
Frame = +1
Query: 223 TIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKN-APPWGKEQTEAIKFLKKAAQNPP 281
++ I+ F G+ ++ R F+P+ S S L+++++KN A WG++Q +A LK+ P
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAP 288
Query: 282 PLKIPGDGKRI-LQTDASDHYWAAVLIEEIDGQKLYCGHASGQFKDAELHYHTTFKETLA 340
L +P K L+ DAS AVL++ G + + S + A L+Y T KE A
Sbjct: 289 VLALPDFSKTFELECDASGVGVRAVLLQ--GGHPI--AYFSEKLHSATLNYPTYDKELYA 456
Query: 341 VKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNK 375
+ + ++ L F + D+ S K + K+K
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSL-KYIRGKSK 558
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 55.5 bits (132), Expect = 4e-08
Identities = 37/133 (27%), Positives = 62/133 (45%), Gaps = 1/133 (0%)
Frame = +2
Query: 83 FSKFDMKSGFWQLGLDPKDRPKTAFCIPNAHYQWTVLPFGLKVAPSLFQKAMTKIFEPIL 142
FS D SG+ Q+ + +D KT F + + V+ FGLK + +Q+AM +F ++
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 143 HTSL-VYIDDVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTE 201
H + VY+DD++ S E H L + + K+ + L+ K K LG ++
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 202 GKFVPQPHIVEEL 214
P V+ L
Sbjct: 362 KGIEIDPEKVKAL 400
>BI317507
Length = 359
Score = 38.5 bits (88), Expect = 0.006
Identities = 24/114 (21%), Positives = 54/114 (47%), Gaps = 1/114 (0%)
Frame = -1
Query: 151 DVLLFSHDEESHKKFLQQFYDICSKHGVMLSSRKSQIATKEVDFLGMHFTEGKFVPQPHI 210
++L++S D +SH L D+ K ++ + +K + +++LG ++ +
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 211 VEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSK-LSKMLRKNAPPWG 263
V+ + ++P +K++ FL + Y R FI K + L+ + + + WG
Sbjct: 173 VKSVIEWP-VPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKWG 15
>BM143397
Length = 408
Score = 32.0 bits (71), Expect = 0.53
Identities = 18/69 (26%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Frame = -3
Query: 345 IKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTPPNPQIL-RLKEWFSLYDFSVKHVQGSK 403
I +F L +++D +L+ K + QI R + S++DF +++ + +
Sbjct: 406 ISKFQSDLLNQKILIRVDCKLAKDILQKDVKNLASKQIFARWQAILSVFDFDIEYFKDTS 227
Query: 404 NLIPDYLSR 412
N +PDYL+R
Sbjct: 226 NSLPDYLTR 200
>TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance
protein-like protein (Fragment), partial (74%)
Length = 781
Score = 31.2 bits (69), Expect = 0.91
Identities = 26/124 (20%), Positives = 53/124 (41%), Gaps = 9/124 (7%)
Frame = +3
Query: 122 GLKVAPSLFQ---KAMTKIFEPILHTSLVYIDDVLLFSHDEESHKKFLQQFYDICSKHGV 178
G ++P LF +T+ + I+ +++ DD++L E + L+ + HG
Sbjct: 144 GSTLSPYLFTLILDVLTEQIQEIVSRCMLFADDIVLLGESREKLNERLETWRRALETHGF 323
Query: 179 MLSSRKSQIATKEVDFLGMHFTEGKFVPQ------PHIVEELPKFPDEGLTIKQIQQFLG 232
LS KS +++ F + + V HI+ ++ +F G I+ + G
Sbjct: 324 RLSRSKS-------EYMECKFNKRRRVSNSEVKIGDHIIPQVTRFKYLGSVIQDDGEIEG 482
Query: 233 ILNY 236
+N+
Sbjct: 483 DVNH 494
>BQ627806
Length = 435
Score = 31.2 bits (69), Expect = 0.91
Identities = 14/49 (28%), Positives = 26/49 (52%)
Frame = +1
Query: 329 LHYHTTFKETLAVKYGIKRFDFHLRGHHFEVQIDNSSFPKMLEFKNKTP 377
L T +E A+ +K++ +L GHHF + D+ S +++ +TP
Sbjct: 220 LRASTYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTP 366
>TC212437 weakly similar to UP|Q9SH20 (Q9SH20) F28K19.1, partial (14%)
Length = 951
Score = 30.0 bits (66), Expect = 2.0
Identities = 18/55 (32%), Positives = 30/55 (53%), Gaps = 5/55 (9%)
Frame = -1
Query: 169 FYDICSKHGVMLSSRKSQIATKEVDFLGMH----FTEGKFVPQP-HIVEELPKFP 218
F D+ + V S + +++ + FL +H + E K VP P H+VE++PK P
Sbjct: 825 FVDVPYQSRVANSWQHCRLSYQYASFLQLHSAIIYAENKQVPPPIHLVEKIPKVP 661
>BI785135
Length = 420
Score = 29.6 bits (65), Expect = 2.6
Identities = 15/59 (25%), Positives = 32/59 (53%)
Frame = +1
Query: 208 PHIVEELPKFPDEGLTIKQIQQFLGILNYIRDFIPHVSKYTSKLSKMLRKNAPPWGKEQ 266
P ++ +P++P +I+ I F + N+ + F+P S + L +++R + P W + Q
Sbjct: 223 PKRIKVIPEWPTPP-SIR*IWGFHNLTNFYKRFVP*FSILVAPLIELVRNHVPSWEEAQ 396
>TC226619 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxide
glutathione peroxidase, complete
Length = 956
Score = 28.9 bits (63), Expect = 4.5
Identities = 13/45 (28%), Positives = 23/45 (50%)
Frame = +3
Query: 365 SFPKMLEFKNKTPPNPQILRLKEWFSLYDFSVKHVQGSKNLIPDY 409
SF + + P P ++ + S+YDF+VK + G+ + DY
Sbjct: 165 SFAFFFFYSHTYPSTPSLMAEQSSNSIYDFTVKDISGNDVSLNDY 299
>TC219992 similar to UP|Q75M40 (Q75M40) Unknow protein, partial (33%)
Length = 937
Score = 28.5 bits (62), Expect = 5.9
Identities = 12/28 (42%), Positives = 15/28 (52%)
Frame = +3
Query: 92 FWQLGLDPKDRPKTAFCIPNAHYQWTVL 119
FW+ P D K CIP+ HY T+L
Sbjct: 45 FWRAHYIPADTSKVHNCIPDEHYVQTLL 128
>TC213097
Length = 429
Score = 28.5 bits (62), Expect = 5.9
Identities = 14/54 (25%), Positives = 25/54 (45%), Gaps = 5/54 (9%)
Frame = -2
Query: 47 KRLVVDYRPLNQFIKDDKF-----PLPKIYSLYSYIKNAHIFSKFDMKSGFWQL 95
KR ++ Y L++ D + P+ + + + H+F K + SGFW L
Sbjct: 338 KRRIMTYTILSRITTDQIYFQ**SPVRYLTETFESTNDRHVFRKATLNSGFWSL 177
>TC219257 weakly similar to UP|Q6PLP6 (Q6PLP6) Cell wall protein GP2
(Fragment), partial (4%)
Length = 933
Score = 28.5 bits (62), Expect = 5.9
Identities = 12/39 (30%), Positives = 23/39 (58%)
Frame = -1
Query: 248 TSKLSKMLRKNAPPWGKEQTEAIKFLKKAAQNPPPLKIP 286
+S S ++ +++ WG Q +++ K A+ PPPL+ P
Sbjct: 753 SSTFSALIGRSSSNWGFLQLLSVRLSGKVARPPPPLEEP 637
>TC216330 similar to UP|Q6PMT1 (Q6PMT1) Methylthioribose kinase , partial
(93%)
Length = 1796
Score = 28.1 bits (61), Expect = 7.7
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Frame = -2
Query: 348 FDFHLRGH-HFEVQIDNSSFPKMLEFKNKTPPNPQIL 383
FD L H H E + SSFP+ML+++N QIL
Sbjct: 1129 FDHLLDQHAHLEQRKPKSSFPRMLQYQNPLVHKMQIL 1019
>TC218873 similar to UP|PSK3_ARATH (Q9M2Y0) Phytosulfokines 3 precursor
(AtPSK3) [Contains: Phytosulfokine-alpha (PSK-alpha)
(Phytosulfokine-a); Phytosulfokine-beta (PSK-beta)
(Phytosulfokine-b)], partial (41%)
Length = 570
Score = 28.1 bits (61), Expect = 7.7
Identities = 11/23 (47%), Positives = 15/23 (64%)
Frame = -2
Query: 353 RGHHFEVQIDNSSFPKMLEFKNK 375
+G H +QI +S+PK E KNK
Sbjct: 524 KGIHLSIQIIENSYPKQQEMKNK 456
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,863,774
Number of Sequences: 63676
Number of extensions: 295531
Number of successful extensions: 1380
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1366
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1372
length of query: 418
length of database: 12,639,632
effective HSP length: 100
effective length of query: 318
effective length of database: 6,272,032
effective search space: 1994506176
effective search space used: 1994506176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0490.11


