

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
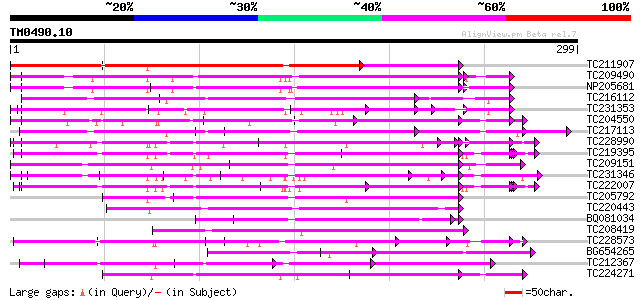
Query= TM0490.10
(299 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211907 weakly similar to UP|Q8S9I3 (Q8S9I3) AT5g20480/F7C8_70,... 196 1e-50
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 114 4e-26
NP205681 receptor protein kinase-like protein 109 2e-24
TC216112 similar to UP|Q8GTD6 (Q8GTD6) Polygalacturonase inhibit... 108 4e-24
TC231353 weakly similar to UP|Q949G9 (Q949G9) HcrVf1 protein, pa... 103 9e-23
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 103 1e-22
TC217113 similar to UP|Q708X5 (Q708X5) Leucine rich repeat prote... 101 4e-22
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 101 5e-22
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 100 1e-21
TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-l... 99 2e-21
TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 98 4e-21
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 97 9e-21
TC205792 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resis... 96 3e-20
TC220443 weakly similar to UP|Q9S711 (Q9S711) ESTs C22657(S0014)... 93 2e-19
BQ081034 92 4e-19
TC208419 weakly similar to UP|Q6X1D9 (Q6X1D9) Resistance protein... 91 6e-19
TC228573 90 1e-18
BG654265 weakly similar to GP|3894385|gb|A Hcr2-0A {Lycopersicon... 90 1e-18
TC212367 weakly similar to UP|Q9ZTJ6 (Q9ZTJ6) Hcr2-5D, partial (... 85 5e-17
TC224271 weakly similar to UP|Q84XU7 (Q84XU7) Receptor-like prot... 84 1e-16
>TC211907 weakly similar to UP|Q8S9I3 (Q8S9I3) AT5g20480/F7C8_70, partial
(5%)
Length = 595
Score = 196 bits (497), Expect = 1e-50
Identities = 102/188 (54%), Positives = 129/188 (68%), Gaps = 1/188 (0%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYL 59
LRN S +T LDLS N I G +P W+W+LN L LN+S NLL++LEGP+Q N S+L L
Sbjct: 46 LRNQSKITTLDLSSNNIQGSIPTWIWQLNSLVQLNLSHNLLSNLEGPVQ--NPSSNLRLL 219
Query: 60 DLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDS 119
DLH+N LQG + IFPV+ Y+DYS N FS IP DIG+++SS IFLSLS N GNIP S
Sbjct: 220 DLHDNHLQGKLQIFPVHASYLDYSSNNFSFTIPSDIGNFLSSTIFLSLSKNNLSGNIPQS 399
Query: 120 LCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLST 179
LC+++ + VLD S N+ +G IP CL + E L VL+L+ N G+IPD FP SC L T
Sbjct: 400 LCSSSSMLVLDFSYNHLNGKIPECL---TQSERLVVLDLQHNKFYGSIPDKFPVSCVLRT 570
Query: 180 LNLRGNQL 187
L+L N L
Sbjct: 571 LDLNSNLL 594
Score = 72.0 bits (175), Expect = 3e-13
Identities = 65/193 (33%), Positives = 91/193 (46%), Gaps = 3/193 (1%)
Frame = +1
Query: 50 LNNVSSLHYLDLHNNQLQGPIP--IFPVN-VVYVDYSRNRFSSVIPQDIGDYMSSAIFLS 106
L N S + LDL +N +QG IP I+ +N +V ++ S N S+ + + + S+ L
Sbjct: 46 LRNQSKITTLDLSSNNIQGSIPTWIWQLNSLVQLNLSHNLLSN-LEGPVQNPSSNLRLLD 222
Query: 107 LSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGT 166
L DN G + +A+ LD S NNFS TIPS + N L T
Sbjct: 223 LHDNHLQGKLQIFPVHAS---YLDYSSNNFSFTIPSDI---------------GNFLSST 348
Query: 167 IPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLR 226
I L+L N L G IP+SL S++ VLD NH+ G P L L
Sbjct: 349 I-----------FLSLSKNNLSGNIPQSLCSSSSMLVLDFSYNHLNGKIPECLTQSERLV 495
Query: 227 VLILRNNRFQGSL 239
VL L++N+F GS+
Sbjct: 496 VLDLQHNKFYGSI 534
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 114 bits (286), Expect = 4e-26
Identities = 95/288 (32%), Positives = 140/288 (47%), Gaps = 28/288 (9%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L L +S+N + GVLP L L +L +LNIS N+ + P Q + ++ L LD+++N
Sbjct: 385 LENLTVSQNNLTGVLPKELAALTSLKHLNISHNVFSG-HFPGQIILPMTKLEVLDVYDNN 561
Query: 66 LQGPIPIFPVNVVYVDYSR---NRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
GP+P+ V + + Y + N FS IP+ ++ S FLSLS N G IP SL
Sbjct: 562 FTGPLPVELVKLEKLKYLKLDGNYFSGSIPESYSEFKSLE-FLSLSTNSLSGKIPKSLSK 738
Query: 123 ATGLQVLDLSINN-FSGTIP---------------SCLMT------MAKPENLEVLNLRD 160
L+ L L NN + G IP SC ++ +A NL+ L L+
Sbjct: 739 LKTLRYLKLGYNNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQI 918
Query: 161 NNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLK 220
NNL GTIP A L +L+L N L G IP S +Q L +++ +N++ G P F+
Sbjct: 919 NNLTGTIPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVG 1098
Query: 221 NISTLRVLILRNNRFQGSL--GCGQVNDEPWKILQIMDIAFNNFSGTL 266
+ L L L +N F L GQ L+ D+ N+F+G +
Sbjct: 1099ELPNLETLQLWDNNFSFVLPPNLGQNGK-----LKFFDVIKNHFTGLI 1227
Score = 108 bits (271), Expect = 2e-24
Identities = 78/248 (31%), Positives = 122/248 (48%), Gaps = 6/248 (2%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLTD--LEGPL-QKLNNVSSLH 57
L + L + D+ KN G++P L + + + ++TD GP+ ++ N SL
Sbjct: 1165 LGQNGKLKFFDVIKNHFTGLIPRDLCKSG----RLQTIMITDNFFRGPIPNEIGNCKSLT 1332
Query: 58 YLDLHNNQLQGPIP--IFPV-NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHG 114
+ NN L G +P IF + +V ++ + NRF+ +P +I S L+LS+N F G
Sbjct: 1333 KIRASNNYLNGVVPSGIFKLPSVTIIELANNRFNGELPPEISG--ESLGILTLSNNLFSG 1506
Query: 115 NIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPAS 174
IP +L N LQ L L N F G IP + + L V+N+ NNL G IP
Sbjct: 1507 KIPPALKNLRALQTLSLDANEFVGEIPGEVFDLPM---LTVVNISGNNLTGPIPTTLTRC 1677
Query: 175 CFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNR 234
L+ ++L N L G IPK + + L + ++ N I+G P ++ + +L L L NN
Sbjct: 1678 VSLTAVDLSRNMLEGKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNN 1857
Query: 235 FQGSLGCG 242
F G + G
Sbjct: 1858 FIGKVPTG 1881
Score = 102 bits (253), Expect = 3e-22
Identities = 82/260 (31%), Positives = 116/260 (44%), Gaps = 27/260 (10%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNN 64
L LD+ N G LP L +L L YL + N + G + + + SL +L L N
Sbjct: 532 LEVLDVYDNNFTGPLPVELVKLEKLKYLKLDGNYFS---GSIPESYSEFKSLEFLSLSTN 702
Query: 65 QLQGPIPIFPVNVVYVDYSR----NRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSL 120
L G IP + + Y + N + IP + G M S +L LS G IP SL
Sbjct: 703 SLSGKIPKSLSKLKTLRYLKLGYNNAYEGGIPPEFGS-MKSLRYLDLSSCNLSGEIPPSL 879
Query: 121 CNATGLQVLDLSINNFSGTIPSCL---------------------MTMAKPENLEVLNLR 159
N T L L L INN +GTIPS L M+ ++ NL ++N
Sbjct: 880 ANLTNLDTLFLQINNLTGTIPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFF 1059
Query: 160 DNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFL 219
NNL+G++P L TL L N +P +L Q L+ D+ KNH TG P L
Sbjct: 1060QNNLRGSVPSFVGELPNLETLQLWDNNFSFVLPPNLGQNGKLKFFDVIKNHFTGLIPRDL 1239
Query: 220 KNISTLRVLILRNNRFQGSL 239
L+ +++ +N F+G +
Sbjct: 1240CKSGRLQTIMITDNFFRGPI 1299
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 109 bits (272), Expect = 2e-24
Identities = 79/249 (31%), Positives = 124/249 (49%), Gaps = 7/249 (2%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLTD--LEGPL-QKLNNVSSLH 57
L + + D++KN G++P L + + + L+TD GP+ ++ N SL
Sbjct: 1075 LGQNGKFKFFDVTKNHFSGLIPRDLCKSG----RLQTFLITDNFFHGPIPNEIANCKSLT 1242
Query: 58 YLDLHNNQLQGPIP--IFPV-NVVYVDYSRNRFSSVIPQDI-GDYMSSAIFLSLSDNKFH 113
+ NN L G +P IF + +V ++ + NRF+ +P +I GD + L+LS+N F
Sbjct: 1243 KIRASNNYLNGAVPSGIFKLPSVTIIELANNRFNGELPPEISGDSLG---ILTLSNNLFT 1413
Query: 114 GNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPA 173
G IP +L N LQ L L N F G IP + + L V+N+ NNL G IP F
Sbjct: 1414 GKIPPALKNLRALQTLSLDTNEFLGEIPGEVFDLPM---LTVVNISGNNLTGPIPTTFTR 1584
Query: 174 SCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNN 233
L+ ++L N L G IPK + + L + ++ N I+G P ++ + +L L L N
Sbjct: 1585 CVSLAAVDLSRNMLDGEIPKGMKNLTDLSIFNVSINQISGSVPDEIRFMLSLTTLDLSYN 1764
Query: 234 RFQGSLGCG 242
F G + G
Sbjct: 1765 NFIGKVPTG 1791
Score = 107 bits (266), Expect = 9e-24
Identities = 90/286 (31%), Positives = 134/286 (46%), Gaps = 26/286 (9%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L L +S+N + G LP L L +L +LNIS N+ + P + + ++ L LD+++N
Sbjct: 295 LENLTISQNNLTGELPKELAALTSLKHLNISHNVFSGYF-PGKIILPMTELEVLDVYDNN 471
Query: 66 LQGPIPIFPVNVVYVDYSR---NRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
G +P V + + Y + N FS IP+ ++ S FLSLS N GNIP SL
Sbjct: 472 FTGSLPEEFVKLEKLKYLKLDGNYFSGSIPESYSEFKSLE-FLSLSTNSLSGNIPKSLSK 648
Query: 123 ATGLQVLDLSINN-FSGTIP---------------SCLMT------MAKPENLEVLNLRD 160
L++L L NN + G IP SC ++ +A NL+ L L+
Sbjct: 649 LKTLRILKLGYNNAYEGGIPPEFGTMESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQM 828
Query: 161 NNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLK 220
NNL GTIP L +L+L N L G IP +Q L +++ N++ G P F+
Sbjct: 829 NNLTGTIPSELSDMVSLMSLDLSFNGLTGEIPTRFSQLKNLTLMNFFHNNLRGSVPSFVG 1008
Query: 221 NISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
+ L L L N F L + +K D+ N+FSG +
Sbjct: 1009ELPNLETLQLWENNFSSELPQNLGQNGKFK---FFDVTKNHFSGLI 1137
Score = 96.3 bits (238), Expect = 2e-20
Identities = 77/261 (29%), Positives = 115/261 (43%), Gaps = 28/261 (10%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNN 64
L LD+ N G LP +L L YL + N + G + + + SL +L L N
Sbjct: 442 LEVLDVYDNNFTGSLPEEFVKLEKLKYLKLDGNYFS---GSIPESYSEFKSLEFLSLSTN 612
Query: 65 QLQGPIP-----IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDS 119
L G IP + + ++ + Y+ N + IP + G M S +L LS G IP S
Sbjct: 613 SLSGNIPKSLSKLKTLRILKLGYN-NAYEGGIPPEFGT-MESLKYLDLSSCNLSGEIPPS 786
Query: 120 LCNATGLQVLDLSINNFSGTIPSCLMTM---------------------AKPENLEVLNL 158
L N L L L +NN +GTIPS L M ++ +NL ++N
Sbjct: 787 LANMRNLDTLFLQMNNLTGTIPSELSDMVSLMSLDLSFNGLTGEIPTRFSQLKNLTLMNF 966
Query: 159 RDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCF 218
NNL+G++P L TL L N +P++L Q + D+ KNH +G P
Sbjct: 967 FHNNLRGSVPSFVGELPNLETLQLWENNFSSELPQNLGQNGKFKFFDVTKNHFSGLIPRD 1146
Query: 219 LKNISTLRVLILRNNRFQGSL 239
L L+ ++ +N F G +
Sbjct: 1147LCKSGRLQTFLITDNFFHGPI 1209
Score = 86.7 bits (213), Expect = 1e-17
Identities = 61/193 (31%), Positives = 98/193 (50%), Gaps = 1/193 (0%)
Frame = +1
Query: 75 VNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSIN 134
+ VV ++ S +P +IG+ + L++S N G +P L T L+ L++S N
Sbjct: 217 LRVVAINVSFVPLFGHVPPEIGE-LDKLENLTISQNNLTGELPKELAALTSLKHLNISHN 393
Query: 135 NFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKS 194
FSG P ++ LEVL++ DNN G++P+ F L L L GN G IP+S
Sbjct: 394 VFSGYFPGKIILPMT--ELEVLDVYDNNFTGSLPEEFVKLEKLKYLKLDGNYFSGSIPES 567
Query: 195 LAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLIL-RNNRFQGSLGCGQVNDEPWKILQ 253
++ +LE L L N ++G P L + TLR+L L NN ++G + + L+
Sbjct: 568 YSEFKSLEFLSLSTNSLSGNIPKSLSKLKTLRILKLGYNNAYEGGI---PPEFGTMESLK 738
Query: 254 IMDIAFNNFSGTL 266
+D++ N SG +
Sbjct: 739 YLDLSSCNLSGEI 777
>TC216112 similar to UP|Q8GTD6 (Q8GTD6) Polygalacturonase inhibitor-like
protein (Fragment), partial (78%)
Length = 1175
Score = 108 bits (269), Expect = 4e-24
Identities = 79/215 (36%), Positives = 110/215 (50%), Gaps = 5/215 (2%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQ-KLNNVSSLHYLDLHNN 64
L +DL N++ G +P + L+ L LN++ NL++ G + L N+SSL +LDL NN
Sbjct: 54 LRIVDLIGNRLSGSIPADIGRLHRLTVLNVADNLIS---GTIPTSLANLSSLMHLDLRNN 224
Query: 65 QLQGPIPIFPVNVVYVD---YSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
GPIP ++ + S NR S IP + A L LS N+ G IP+SL
Sbjct: 225 LFSGPIPRNFGSLSMLSRALLSGNRLSGAIPSSVSQIYRLAD-LDLSRNQISGPIPESLG 401
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
L L+L +N SG IP L + + LNL N L+G IPD F + + L+
Sbjct: 402 KMAVLSTLNLDMNKLSGPIPVSLFS----SGISDLNLSRNALEGNIPDAFGVRSYFTALD 569
Query: 182 LRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP 216
L N L G IPKS++ S + LDL NH+ G P
Sbjct: 570 LSYNNLKGAIPKSISSASYIGHLDLSHNHLCGKIP 674
Score = 84.0 bits (206), Expect = 8e-17
Identities = 63/190 (33%), Positives = 93/190 (48%), Gaps = 3/190 (1%)
Frame = +3
Query: 80 VDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGT 139
VD NR S IP DIG + L+++DN G IP SL N + L LDL N FSG
Sbjct: 63 VDLIGNRLSGSIPADIGRLHRLTV-LNVADNLISGTIPTSLANLSSLMHLDLRNNLFSGP 239
Query: 140 IPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCS 199
IP +++ L L N L G IP L+ L+L NQ+ GPIP+SL + +
Sbjct: 240 IPRNFGSLSM---LSRALLSGNRLSGAIPSSVSQIYRLADLDLSRNQISGPIPESLGKMA 410
Query: 200 TLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKI---LQIMD 256
L L+L N ++G P L + S + L L N +G++ + + + +D
Sbjct: 411 VLSTLNLDMNKLSGPIPVSLFS-SGISDLNLSRNALEGNI------PDAFGVRSYFTALD 569
Query: 257 IAFNNFSGTL 266
+++NN G +
Sbjct: 570 LSYNNLKGAI 599
>TC231353 weakly similar to UP|Q949G9 (Q949G9) HcrVf1 protein, partial (16%)
Length = 1277
Score = 103 bits (257), Expect = 9e-23
Identities = 81/290 (27%), Positives = 128/290 (43%), Gaps = 54/290 (18%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPN-WLWELNLHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNN 64
L +L+L+ N + G +P+ W+ +L +N+ SN G L Q + +++ L L + NN
Sbjct: 36 LEFLNLASNNLSGEIPDCWMNWTSLVDVNLQSNHFV---GNLPQSMGSLAELQSLQIRNN 206
Query: 65 QLQGPIPIFPVNV------VYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPD 118
L G IFP ++ + +D N S IP +G+ + + L L N F G+IP+
Sbjct: 207 TLSG---IFPTSLKKNNQLISLDLGENNLSGTIPTWVGENLLNVKILRLRSNSFAGHIPN 377
Query: 119 SLCNATGLQVLDLSINNFSGTIPSCL-----MTMAKPEN--------------------- 152
+C + LQVLDL+ NN SG IPSC MT+
Sbjct: 378 EICQMSHLQVLDLAQNNLSGNIPSCFSNLSAMTLKNQSTDPRIYSQAQIYGRYYSSRQSI 557
Query: 153 --------------------LEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIP 192
+ ++L N L G IP L+ LN+ NQL G IP
Sbjct: 558 VSVLLWLKGRGDEYRNILGLVTSIDLSSNKLLGEIPREITYLNGLNFLNMSHNQLIGHIP 737
Query: 193 KSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCG 242
+ + +L+ +D +N + G P + N+S L +L L N +G++ G
Sbjct: 738 QGIGNMRSLQSIDFSRNQLFGEIPPSIANLSFLSMLDLSYNHLKGNIPTG 887
Score = 82.4 bits (202), Expect = 2e-16
Identities = 52/153 (33%), Positives = 79/153 (50%), Gaps = 1/153 (0%)
Frame = +3
Query: 74 PVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSI 133
P+ + +++ + N S IP D +S + ++L N F GN+P S+ + LQ L +
Sbjct: 27 PMLLEFLNLASNNLSGEIP-DCWMNWTSLVDVNLQSNHFVGNLPQSMGSLAELQSLQIRN 203
Query: 134 NNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCF-LSTLNLRGNQLHGPIP 192
N SG P+ L K L L+L +NNL GTIP + + L LR N G IP
Sbjct: 204 NTLSGIFPTSLK---KNNQLISLDLGENNLSGTIPTWVGENLLNVKILRLRSNSFAGHIP 374
Query: 193 KSLAQCSTLEVLDLGKNHITGGFPCFLKNISTL 225
+ Q S L+VLDL +N+++G P N+S +
Sbjct: 375 NEICQMSHLQVLDLAQNNLSGNIPSCFSNLSAM 473
Score = 79.0 bits (193), Expect = 2e-15
Identities = 74/225 (32%), Positives = 100/225 (43%), Gaps = 9/225 (4%)
Frame = +3
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWE--LNLHYLNISSNLLTDLEGPL-QKLNNVSSLH 57
L+ ++ L LDL +N + G +P W+ E LN+ L + SN G + ++ +S L
Sbjct: 234 LKKNNQLISLDLGENNLSGTIPTWVGENLLNVKILRLRSN---SFAGHIPNEICQMSHLQ 404
Query: 58 YLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVI---PQDIGDYMSS--AIFLSLSDNKF 112
LDL N L G IP N+ + I Q G Y SS +I L K
Sbjct: 405 VLDLAQNNLSGNIPSCFSNLSAMTLKNQSTDPRIYSQAQIYGRYYSSRQSIVSVLLWLKG 584
Query: 113 HGNIPDSLCNATGLQV-LDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMF 171
G D N GL +DLS N G IP + + L LN+ N L G IP
Sbjct: 585 RG---DEYRNILGLVTSIDLSSNKLLGEIPREITYL---NGLNFLNMSHNQLIGHIPQGI 746
Query: 172 PASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP 216
L +++ NQL G IP S+A S L +LDL NH+ G P
Sbjct: 747 GNMRSLQSIDFSRNQLFGEIPPSIANLSFLSMLDLSYNHLKGNIP 881
Score = 65.5 bits (158), Expect = 3e-11
Identities = 49/145 (33%), Positives = 74/145 (50%), Gaps = 27/145 (18%)
Frame = +3
Query: 149 KPENLEVLNLRDNNLKGTIPDMF--------------------PAS----CFLSTLNLRG 184
KP LE LNL NNL G IPD + P S L +L +R
Sbjct: 24 KPMLLEFLNLASNNLSGEIPDCWMNWTSLVDVNLQSNHFVGNLPQSMGSLAELQSLQIRN 203
Query: 185 NQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFL-KNISTLRVLILRNNRFQGSLGCGQ 243
N L G P SL + + L LDLG+N+++G P ++ +N+ +++L LR+N F G
Sbjct: 204 NTLSGIFPTSLKKNNQLISLDLGENNLSGTIPTWVGENLLNVKILRLRSNSF-----AGH 368
Query: 244 VNDEPWKI--LQIMDIAFNNFSGTL 266
+ +E ++ LQ++D+A NN SG +
Sbjct: 369 IPNEICQMSHLQVLDLAQNNLSGNI 443
Score = 62.0 bits (149), Expect = 3e-10
Identities = 58/198 (29%), Positives = 95/198 (47%), Gaps = 12/198 (6%)
Frame = +3
Query: 5 STLTYLDLSKNQIHGVLPNWLWELNLHYL-NISSNLLTDLEGPL------QKLNNVSSLH 57
S L LDL++N + G +P+ L+ L N S++ + + + + VS L
Sbjct: 393 SHLQVLDLAQNNLSGNIPSCFSNLSAMTLKNQSTDPRIYSQAQIYGRYYSSRQSIVSVLL 572
Query: 58 YLDLHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIP 117
+L ++ + + + V +D S N+ IP++I Y++ FL++S N+ G+IP
Sbjct: 573 WLKGRGDEYRNILGL----VTSIDLSSNKLLGEIPREI-TYLNGLNFLNMSHNQLIGHIP 737
Query: 118 DSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIP-----DMFP 172
+ N LQ +D S N G IP + ++ L +L+L N+LKG IP F
Sbjct: 738 QGIGNMRSLQSIDFSRNQLFGEIPPSIANLSF---LSMLDLSYNHLKGNIPTGTQLQTFD 908
Query: 173 ASCFLSTLNLRGNQLHGP 190
AS F+ GN L GP
Sbjct: 909 ASSFI------GNNLCGP 944
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 103 bits (256), Expect = 1e-22
Identities = 88/269 (32%), Positives = 132/269 (48%), Gaps = 9/269 (3%)
Frame = +2
Query: 7 LTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L L L NQI G +P+ L L NL + + +N LT PL L L LDL NN
Sbjct: 515 LRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTG-SIPLS-LGFCPLLQSLDLSNNL 688
Query: 66 LQGPIPIFPVN---VVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
L G IP N + +++ S N FS +P + + S FLSL +N G++P+S
Sbjct: 689 LTGAIPYSLANSTKLYWLNLSFNSFSGPLPASL-THSFSLTFLSLQNNNLSGSLPNSWGG 865
Query: 123 ATG-----LQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFL 177
+ LQ L L N F+G +P+ L ++ + L ++L N G IP+ L
Sbjct: 866 NSKNGFFRLQNLILDHNFFTGDVPASLGSLRE---LNEISLSHNKFSGAIPNEIGTLSRL 1036
Query: 178 STLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQG 237
TL++ N L+G +P +L+ S+L +L+ N + P L + L VLIL N+F G
Sbjct: 1037KTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLILSRNQFSG 1216
Query: 238 SLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
+ N L+ +D++ NNFSG +
Sbjct: 1217HIPSSIANISS---LRQLDLSLNNFSGEI 1294
Score = 83.6 bits (205), Expect = 1e-16
Identities = 68/220 (30%), Positives = 106/220 (47%), Gaps = 36/220 (16%)
Frame = +2
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELN-------LHYLNISSNLLT-DLE---GPLQK 49
L + +LT+L L N + G LPN W N L L + N T D+ G L++
Sbjct: 785 LTHSFSLTFLSLQNNNLSGSLPN-SWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRE 961
Query: 50 LNNVSSLHY------------------LDLHNNQLQGPIPIFPVNV---VYVDYSRNRFS 88
LN +S H LD+ NN L G +P N+ ++ N
Sbjct: 962 LNEISLSHNKFSGAIPNEIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLD 1141
Query: 89 SVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMA 148
+ IPQ +G + ++ + LS N+F G+IP S+ N + L+ LDLS+NNFSG IP ++
Sbjct: 1142NQIPQSLGRLRNLSVLI-LSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIP---VSFD 1309
Query: 149 KPENLEVLNLRDNNLKGTIPDM----FPASCFLSTLNLRG 184
+L + N+ N+L G++P + F +S F+ + L G
Sbjct: 1310SQRSLNLFNVSYNSLSGSVPPLLAKKFNSSSFVGNIQLCG 1429
Score = 68.6 bits (166), Expect = 3e-12
Identities = 49/137 (35%), Positives = 65/137 (46%)
Frame = +2
Query: 103 IFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNN 162
I + L G I D + GL+ L L N G+IPS T+ NL + L +N
Sbjct: 446 IVIQLPWKGLRGRITDKIGQLQGLRKLSLHDNQIGGSIPS---TLGLLPNLRGVQLFNNR 616
Query: 163 LKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNI 222
L G+IP L +L+L N L G IP SLA + L L+L N +G P L +
Sbjct: 617 LTGSIPLSLGFCPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHS 796
Query: 223 STLRVLILRNNRFQGSL 239
+L L L+NN GSL
Sbjct: 797 FSLTFLSLQNNNLSGSL 847
Score = 60.5 bits (145), Expect = 9e-10
Identities = 42/123 (34%), Positives = 56/123 (45%)
Frame = +2
Query: 151 ENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNH 210
+ L L+L DN + G+IP L + L N+L G IP SL C L+ LDL N
Sbjct: 509 QGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFCPLLQSLDLSNNL 688
Query: 211 ITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKY 270
+TG P L N + L L L N F G L + L + + NN SG+L +
Sbjct: 689 LTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLTFLSLQN---NNLSGSLPNSW 859
Query: 271 FKN 273
N
Sbjct: 860 GGN 868
>TC217113 similar to UP|Q708X5 (Q708X5) Leucine rich repeat protein
precursor, partial (96%)
Length = 1553
Score = 101 bits (252), Expect = 4e-22
Identities = 74/216 (34%), Positives = 111/216 (51%), Gaps = 5/216 (2%)
Frame = +3
Query: 6 TLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQ-KLNNVSSLHYLDLHN 63
+L LDL N++ G +P + +L+ L LN++ N L+ G + + + SL +LDL N
Sbjct: 396 SLRILDLIGNKLSGEIPADVGKLSRLTVLNLADNALS---GKIPASITQLGSLKHLDLSN 566
Query: 64 NQLQGPIPIFPVNVVYVD---YSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSL 120
NQL G IP N+ + SRN+ + IP + A L LS N+ G++P L
Sbjct: 567 NQLCGEIPEDFGNLGMLSRMLLSRNQLTGKIPVSVSKIYRLAD-LDLSANRLSGSVPFEL 743
Query: 121 CNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTL 180
L L+L N+ G IPS L++ + +LNL N +G+IPD+F + + L
Sbjct: 744 GTMPVLSTLNLDSNSLEGLIPSSLLSNG---GMGILNLSRNGFEGSIPDVFGSHSYFMAL 914
Query: 181 NLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP 216
+L N L G +P SLA + LDL NH+ G P
Sbjct: 915 DLSFNNLKGRVPSSLASAKFIGHLDLSHNHLCGSIP 1022
Score = 75.5 bits (184), Expect = 3e-14
Identities = 58/203 (28%), Positives = 99/203 (48%), Gaps = 5/203 (2%)
Frame = +3
Query: 99 MSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNL 158
+ S L L NK G IP + + L VL+L+ N SG IP+ + + +L+ L+L
Sbjct: 390 LPSLRILDLIGNKLSGEIPADVGKLSRLTVLNLADNALSGKIPASITQLG---SLKHLDL 560
Query: 159 RDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCF 218
+N L G IP+ F LS + L NQL G IP S+++ L LDL N ++G P
Sbjct: 561 SNNQLCGEIPEDFGNLGMLSRMLLSRNQLTGKIPVSVSKIYRLADLDLSANRLSGSVPFE 740
Query: 219 LKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL-----KGKYFKN 273
L + L L L +N +G + +++ + I++++ N F G++ YF
Sbjct: 741 LGTMPVLSTLNLDSNSLEGLIPSSLLSNGG---MGILNLSRNGFEGSIPDVFGSHSYFMA 911
Query: 274 WETMMHDEEDQYVSNFIHKEFLG 296
+ ++ + + S+ +F+G
Sbjct: 912 LDLSFNNLKGRVPSSLASAKFIG 980
Score = 70.9 bits (172), Expect = 7e-13
Identities = 52/160 (32%), Positives = 72/160 (44%)
Frame = +3
Query: 114 GNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPA 173
G IP + L++LDL N SG IP+ + K L VLNL DN L G IP
Sbjct: 363 GEIPTCVTALPSLRILDLIGNKLSGEIPA---DVGKLSRLTVLNLADNALSGKIPASITQ 533
Query: 174 SCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNN 233
L L+L NQL G IP+ L + L +N +TG P + I L L L N
Sbjct: 534 LGSLKHLDLSNNQLCGEIPEDFGNLGMLSRMLLSRNQLTGKIPVSVSKIYRLADLDLSAN 713
Query: 234 RFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKN 273
R GS+ ++ P +L +++ N+ G + N
Sbjct: 714 RLSGSVPF-ELGTMP--VLSTLNLDSNSLEGLIPSSLLSN 824
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 101 bits (251), Expect = 5e-22
Identities = 84/290 (28%), Positives = 141/290 (47%), Gaps = 30/290 (10%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L++L L+ N+ G +P L+ L +LN+S+N+ + P Q LN +++L LDL+NN
Sbjct: 361 LSHLSLADNKFSGPIPASFSALSALRFLNLSNNVF-NATFPSQ-LNRLANLEVLDLYNNN 534
Query: 66 LQGPIPI----FPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNK---------- 111
+ G +P+ P+ + ++ N FS IP + G + +L+LS N+
Sbjct: 535 MTGELPLSVAAMPL-LRHLHLGGNFFSGQIPPEYGTWQHLQ-YLALSGNELAGTIAPELG 708
Query: 112 ---------------FHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVL 156
+ G IP + N + L LD + SG IP+ + K +NL+ L
Sbjct: 709 NLSSLRELYIGYYNTYSGGIPPEIGNLSNLVRLDAAYCGLSGEIPA---ELGKLQNLDTL 879
Query: 157 NLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP 216
L+ N L G++ + L +++L N L G +P S A+ L +L+L +N + G P
Sbjct: 880 FLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIP 1059
Query: 217 CFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
F+ + L VL L N F GS+ N+ L ++D++ N +GTL
Sbjct: 1060EFVGELPALEVLQLWENNFTGSIPQNLGNNGR---LTLVDLSSNKITGTL 1200
Score = 96.3 bits (238), Expect = 2e-20
Identities = 76/251 (30%), Positives = 121/251 (47%), Gaps = 8/251 (3%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLT---DLEGPL-QKLNNVSSL 56
L N+ LT +DLS N+I G LP N+ Y N L+T L GP+ L SL
Sbjct: 1138 LGNNGRLTLVDLSSNKITGTLPP-----NMCYGNRLQTLITLGNYLFGPIPDSLGKCKSL 1302
Query: 57 HYLDLHNNQLQGPIP--IFPV-NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFH 113
+ + + N L G IP +F + + V+ N + P+D G + +SLS+N+
Sbjct: 1303 NRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPED-GSIATDLGQISLSNNQLS 1479
Query: 114 GNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPA 173
G++P ++ N T +Q L L+ N F+G IP + + + L ++ N G I
Sbjct: 1480 GSLPSTIGNFTSMQKLLLNGNEFTGRIPPQIGML---QQLSKIDFSHNKFSGPIAPEISK 1650
Query: 174 SCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNN 233
L+ ++L GN+L G IP + L L+L +NH+ G P + ++ +L + N
Sbjct: 1651 CKLLTFIDLSGNELSGEIPNKITSMRILNYLNLSRNHLDGSIPGNIASMQSLTSVDFSYN 1830
Query: 234 RFQGSL-GCGQ 243
F G + G GQ
Sbjct: 1831 NFSGLVPGTGQ 1863
Score = 93.2 bits (230), Expect = 1e-19
Identities = 76/239 (31%), Positives = 113/239 (46%), Gaps = 4/239 (1%)
Frame = +1
Query: 3 NHSTLTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDL 61
N S L LD + + G +P L +L NL L + N L+ P +L ++ SL +DL
Sbjct: 784 NLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTP--ELGSLKSLKSMDL 957
Query: 62 HNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
NN L G +P F+ + + L+L NK HG IP+ +
Sbjct: 958 SNNMLSGEVPA-------------SFAE---------LKNLTLLNLFRNKLHGAIPEFVG 1071
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCF---LS 178
L+VL L NNF+G+IP L + L +++L N + GT+P P C+ L
Sbjct: 1072ELPALEVLQLWENNFTGSIPQNLGNNGR---LTLVDLSSNKITGTLP---PNMCYGNRLQ 1233
Query: 179 TLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQG 237
TL GN L GPIP SL +C +L + +G+N + G P L + L + L++N G
Sbjct: 1234TLITLGNYLFGPIPDSLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTG 1410
Score = 92.4 bits (228), Expect = 2e-19
Identities = 75/245 (30%), Positives = 120/245 (48%), Gaps = 6/245 (2%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNL-HYLNISSNLLTDLEGPLQKLNNVSSLHYL 59
L + L LDL N + G LP + + L +L++ N + P + L YL
Sbjct: 487 LNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQIPP--EYGTWQHLQYL 660
Query: 60 DLHNNQLQGPIP-----IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHG 114
L N+L G I + + +Y+ Y N +S IP +IG+ +S+ + L + G
Sbjct: 661 ALSGNELAGTIAPELGNLSSLRELYIGYY-NTYSGGIPPEIGN-LSNLVRLDAAYCGLSG 834
Query: 115 NIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPAS 174
IP L L L L +N SG++ L ++ ++L+ ++L +N L G +P F
Sbjct: 835 EIPAELGKLQNLDTLFLQVNALSGSLTPELGSL---KSLKSMDLSNNMLSGEVPASFAEL 1005
Query: 175 CFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNR 234
L+ LNL N+LHG IP+ + + LEVL L +N+ TG P L N L ++ L +N+
Sbjct: 1006KNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQNLGNNGRLTLVDLSSNK 1185
Query: 235 FQGSL 239
G+L
Sbjct: 1186ITGTL 1200
Score = 82.0 bits (201), Expect = 3e-16
Identities = 76/234 (32%), Positives = 111/234 (46%), Gaps = 12/234 (5%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPN--WLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDL-HN 63
L +L L N G +P W+ +L YL +S N L P +L N+SSL L + +
Sbjct: 577 LRHLHLGGNFFSGQIPPEYGTWQ-HLQYLALSGNELAGTIAP--ELGNLSSLRELYIGYY 747
Query: 64 NQLQGPIPIFPV-----NVVYVDYSRNRFSSVIPQDIGDYMS-SAIFLSLSDNKFHGNIP 117
N G IP P N+V +D + S IP ++G + +FL + N G++
Sbjct: 748 NTYSGGIP--PEIGNLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQV--NALSGSLT 915
Query: 118 DSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFL 177
L + L+ +DLS N SG +P+ + A+ +NL +LNL N L G IP+ L
Sbjct: 916 PELGSLKSLKSMDLSNNMLSGEVPA---SFAELKNLTLLNLFRNKLHGAIPEFVGELPAL 1086
Query: 178 STLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFP---CFLKNISTLRVL 228
L L N G IP++L L ++DL N ITG P C+ + TL L
Sbjct: 1087EVLQLWENNFTGSIPQNLGNNGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITL 1248
Score = 63.9 bits (154), Expect = 8e-11
Identities = 44/151 (29%), Positives = 73/151 (48%), Gaps = 3/151 (1%)
Frame = +1
Query: 132 SINNFSGTIPSCL---MTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLH 188
++++++ + P C +T ++ LNL +L GT+ D FLS L+L N+
Sbjct: 217 ALSSWNSSTPFCSWFGLTCDSRRHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFS 396
Query: 189 GPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEP 248
GPIP S + S L L+L N FP L ++ L VL L NN G L ++
Sbjct: 397 GPIPASFSALSALRFLNLSNNVFNATFPSQLNRLANLEVLDLYNNNMTGEL---PLSVAA 567
Query: 249 WKILQIMDIAFNNFSGTLKGKYFKNWETMMH 279
+L+ + + N FSG + +Y W+ + +
Sbjct: 568 MPLLRHLHLGGNFFSGQIPPEY-GTWQHLQY 657
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 99.8 bits (247), Expect = 1e-21
Identities = 83/274 (30%), Positives = 124/274 (44%), Gaps = 9/274 (3%)
Frame = +3
Query: 3 NHSTLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYLDL 61
N S L LD + + G +P L +L L L + N L+ P +L N+ SL +DL
Sbjct: 468 NLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTP--ELGNLKSLKSMDL 641
Query: 62 HNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
NN L G IP RF + + L+L NK HG IP+ +
Sbjct: 642 SNNMLSGEIPA-------------RFGE---------LKNITLLNLFRNKLHGAIPEFIG 755
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
L+V+ L NNF+G+IP L K L +++L N L GT+P + L TL
Sbjct: 756 ELPALEVVQLWENNFTGSIPEGL---GKNGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLI 926
Query: 182 LRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQG---- 237
GN L GPIP+SL C +L + +G+N + G P L + L + L++N G
Sbjct: 927 TLGNFLFGPIPESLGSCESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPE 1106
Query: 238 ----SLGCGQVNDEPWKILQIMDIAFNNFSGTLK 267
++ GQ+ ++ ++ + NFS K
Sbjct: 1107VGSVAVNLGQITLSNNQLSGVLPPSIGNFSSVQK 1208
Score = 95.1 bits (235), Expect = 3e-20
Identities = 73/239 (30%), Positives = 121/239 (50%), Gaps = 6/239 (2%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L LDL N + GVLP + ++ NL +L++ N + P + L YL + N+
Sbjct: 189 LEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPP--EYGRWQRLQYLAVSGNE 362
Query: 66 LQGPIP-----IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSL 120
L+G IP + + +Y+ Y N ++ IP +IG+ +S + L + G IP +L
Sbjct: 363 LEGTIPPEIGNLSSLRELYIGYY-NTYTGGIPPEIGN-LSELVRLDAAYCGLSGEIPAAL 536
Query: 121 CNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTL 180
L L L +N SG++ L + ++L+ ++L +N L G IP F ++ L
Sbjct: 537 GKLQKLDTLFLQVNALSGSLTPELGNL---KSLKSMDLSNNMLSGEIPARFGELKNITLL 707
Query: 181 NLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
NL N+LHG IP+ + + LEV+ L +N+ TG P L L ++ L +N+ G+L
Sbjct: 708 NLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGLGKNGRLNLVDLSSNKLTGTL 884
Score = 94.7 bits (234), Expect = 4e-20
Identities = 77/288 (26%), Positives = 136/288 (46%), Gaps = 28/288 (9%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L+ L L+ N+ G +P L L+ L +LN+S+N+ + +L+ + +L LDL+NN
Sbjct: 45 LSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFP--SELSRLQNLEVLDLYNNN 218
Query: 66 LQGPIPIFPV---NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
+ G +P+ N+ ++ N FS IP + G + +L++S N+ G IP + N
Sbjct: 219 MTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQ-YLAVSGNELEGTIPPEIGN 395
Query: 123 ATGLQVLDLS-INNFSGTIPSCL---------------------MTMAKPENLEVLNLRD 160
+ L+ L + N ++G IP + + K + L+ L L+
Sbjct: 396 LSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQV 575
Query: 161 NNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLK 220
N L G++ L +++L N L G IP + + +L+L +N + G P F+
Sbjct: 576 NALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIG 755
Query: 221 NISTLRVLILRNNRFQGSL--GCGQVNDEPWKILQIMDIAFNNFSGTL 266
+ L V+ L N F GS+ G G+ L ++D++ N +GTL
Sbjct: 756 ELPALEVVQLWENNFTGSIPEGLGKNGR-----LNLVDLSSNKLTGTL 884
Score = 82.8 bits (203), Expect = 2e-16
Identities = 74/290 (25%), Positives = 127/290 (43%), Gaps = 28/290 (9%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQK-LNNVSSLHYLDLHNN 64
+T L+L +N++HG +P ++ EL L + + N T G + + L L+ +DL +N
Sbjct: 696 ITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFT---GSIPEGLGKNGRLNLVDLSSN 866
Query: 65 QLQGPIPIFPV--NVVYVDYSRNRFS-SVIPQDIG------------DYMSSAI------ 103
+L G +P + N + + F IP+ +G ++++ +I
Sbjct: 867 KLTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMGENFLNGSIPRGLFG 1046
Query: 104 -----FLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNL 158
+ L DN G P+ A L + LS N SG +P + + +++ L L
Sbjct: 1047LPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGVLPPSIGNFS---SVQKLIL 1217
Query: 159 RDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCF 218
N G IP LS ++ GN+ GPI ++QC L LDL +N ++G P
Sbjct: 1218DGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNE 1397
Query: 219 LKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKG 268
+ + L L L N G + + + L +D ++NN SG + G
Sbjct: 1398ITGMRILNYLNLSRNHLVGGI---PSSISSMQSLTSVDFSYNNLSGLVPG 1538
Score = 52.4 bits (124), Expect = 2e-07
Identities = 35/104 (33%), Positives = 50/104 (47%)
Frame = +3
Query: 176 FLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRF 235
FLS L+L N+ GPIP SL+ S L L+L N FP L + L VL L NN
Sbjct: 42 FLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRLQNLEVLDLYNNNM 221
Query: 236 QGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKNWETMMH 279
G L + L+ + + N FSG + +Y + W+ + +
Sbjct: 222 TGVLPLAVAQ---MQNLRHLHLGGNFFSGQIPPEYGR-WQRLQY 341
>TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-like, partial
(31%)
Length = 1329
Score = 99.0 bits (245), Expect = 2e-21
Identities = 98/326 (30%), Positives = 145/326 (44%), Gaps = 54/326 (16%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQK-LNNVSSLHY 58
L N LT LDLS N G +P +L NL L++ N D+ G + + + + SL
Sbjct: 10 LSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYN---DMSGTVPEGIAQLPSLET 180
Query: 59 LDLHNNQLQGPIPIF---PVNVVYVDYSRNRFSSVIPQDI---GDYM------------- 99
L + NN+ G +P + +VD S N IP DI G+
Sbjct: 181 LLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGGL 360
Query: 100 ------SSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENL 153
SS + L L DN F G I + +DLS NNF G IPS +++ L
Sbjct: 361 SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPS---DISQATQL 531
Query: 154 EVLNLRDNN-LKGTIPD-----------------------MFPASCFLSTLNLRGNQLHG 189
E N+ N L G IP +F + +S ++L N L G
Sbjct: 532 EYFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPLFESCKSISVIDLDSNSLSG 711
Query: 190 PIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGC--GQVNDE 247
IP +++C LE ++L N++TG P L +I L V+ L NN+F G + G ++
Sbjct: 712 TIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSN- 888
Query: 248 PWKILQIMDIAFNNFSGTL-KGKYFK 272
LQ+++++FNN SG++ GK FK
Sbjct: 889 ----LQLLNVSFNNISGSIPTGKSFK 954
Score = 67.8 bits (164), Expect = 6e-12
Identities = 43/123 (34%), Positives = 64/123 (51%)
Frame = +1
Query: 117 PDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCF 176
P L N L LDLS N F+G+IP + ENL +L++ N++ GT+P+
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDL---ENLRLLSVMYNDMSGTVPEGIAQLPS 171
Query: 177 LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQ 236
L TL + N+ G +P+SL + S L+ +D N + G P + L LIL +N+F
Sbjct: 172 LETLLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFT 351
Query: 237 GSL 239
G L
Sbjct: 352 GGL 360
>TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial (14%)
Length = 1790
Score = 98.2 bits (243), Expect = 4e-21
Identities = 79/239 (33%), Positives = 114/239 (47%), Gaps = 26/239 (10%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPL-QKLNNVSSLHY 58
L+N ST+ ++D+ NQ+ +P+W+WE+ L L + SN + G + QK+ +SSL
Sbjct: 1051 LQNCSTMKFIDMGNNQLSDTIPDWMWEMQYLMVLRLRSN---NFNGSITQKMCQLSSLIV 1221
Query: 59 LDLHNNQLQGPIP----------------IFPVNVVY-VDYSRNRFSSVI----PQDIGD 97
LD NN L G IP P + Y D+S N + + +D +
Sbjct: 1222 LDHGNNSLSGSIPNCLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLVPKKDELE 1401
Query: 98 YMSSAIF---LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLE 154
Y + I + LS NK G IP + + L+ L+LS N+ SG IP+ M K + LE
Sbjct: 1402 YRDNLILVRMIDLSSNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPN---DMGKMKLLE 1572
Query: 155 VLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITG 213
L+L NN+ G IP FLS LNL + L G IP S S E+ G + G
Sbjct: 1573 SLDLSLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRIPTSTQLQSFEELSYTGNPELCG 1749
Score = 97.8 bits (242), Expect = 5e-21
Identities = 72/243 (29%), Positives = 113/243 (45%), Gaps = 4/243 (1%)
Frame = +1
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLD 60
L+ S++ L +SK I ++P+W W L + +LD
Sbjct: 550 LKRQSSVKVLTMSKAGIADLVPSWFWIWTLQ------------------------IEFLD 657
Query: 61 LHNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSL 120
L NN L+G + +N ++ S N F +P ++ L++++N G I L
Sbjct: 658 LSNNLLRGDLSNIFLNSSVINLSSNLFKGRLPS----VSANVEVLNVANNSISGTISPFL 825
Query: 121 C---NATG-LQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCF 176
C NAT L VLD S N SG + C + + L +NL NN+ G IP+
Sbjct: 826 CGKPNATNKLSVLDFSNNVLSGDLGHCWVHW---QALVHVNLGSNNMSGEIPNSMGYLSQ 996
Query: 177 LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQ 236
L +L L N+ G IP +L CST++ +D+G N ++ P ++ + L VL LR+N F
Sbjct: 997 LESLLLDDNRFSGYIPSTLQNCSTMKFIDMGNNQLSDTIPDWMWEMQYLMVLRLRSNNFN 1176
Query: 237 GSL 239
GS+
Sbjct: 1177GSI 1185
Score = 91.3 bits (225), Expect = 5e-19
Identities = 82/281 (29%), Positives = 122/281 (43%), Gaps = 48/281 (17%)
Frame = +1
Query: 7 LTYLDLSKNQIHGVLPN-WLWELNLHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNN 64
L+ LD S N + G L + W+ L ++N+ SN ++ G + + +S L L L +N
Sbjct: 853 LSVLDFSNNVLSGDLGHCWVHWQALVHVNLGSN---NMSGEIPNSMGYLSQLESLLLDDN 1023
Query: 65 QLQGPIPIFPVN---VVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
+ G IP N + ++D N+ S IP + + M + L L N F+G+I +C
Sbjct: 1024 RFSGYIPSTLQNCSTMKFIDMGNNQLSDTIPDWMWE-MQYLMVLRLRSNNFNGSITQKMC 1200
Query: 122 NATGLQVLDLSINNFSGTIPSCL---MTMA------------------------------ 148
+ L VLD N+ SG+IP+CL TMA
Sbjct: 1201 QLSSLIVLDHGNNSLSGSIPNCLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLV 1380
Query: 149 -KPENLE---------VLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQC 198
K + LE +++L N L G IP L LNL N L G IP + +
Sbjct: 1381 PKKDELEYRDNLILVRMIDLSSNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKM 1560
Query: 199 STLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
LE LDL N+I+G P L ++S L L L + G +
Sbjct: 1561 KLLESLDLSLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRI 1683
Score = 81.3 bits (199), Expect = 5e-16
Identities = 49/128 (38%), Positives = 69/128 (53%)
Frame = +1
Query: 112 FHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMF 171
+ G IP + + ++ LDL N SG +P L + ++LEVL+L +N IP F
Sbjct: 19 YRGKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQL---KHLEVLDLSNNTFTCPIPSPF 189
Query: 172 PASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILR 231
L TLNL N+L+G IPKS L+VL+LG N +TG P L +S L L L
Sbjct: 190 ANLSSLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLS 369
Query: 232 NNRFQGSL 239
+N +GS+
Sbjct: 370 SNLLEGSI 393
Score = 75.9 bits (185), Expect = 2e-14
Identities = 52/132 (39%), Positives = 65/132 (48%)
Frame = +1
Query: 82 YSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIP 141
Y+ + IPQ I + + L L +N+ G +PDSL L+VLDLS N F+ IP
Sbjct: 4 YTVTFYRGKIPQIISS-LQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIP 180
Query: 142 SCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTL 201
S A +L LNL N L GTIP F L LNL N L G +P +L S L
Sbjct: 181 S---PFANLSSLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNL 351
Query: 202 EVLDLGKNHITG 213
LDL N + G
Sbjct: 352 VTLDLSSNLLEG 387
Score = 75.9 bits (185), Expect = 2e-14
Identities = 61/187 (32%), Positives = 95/187 (50%), Gaps = 4/187 (2%)
Frame = +1
Query: 48 QKLNNVSSLHYLDLHNNQLQGPIPIFPVNVVY---VDYSRNRFSSVIPQDIGDYMSSAIF 104
Q ++++ ++ LDL NNQL GP+P + + +D S N F+ IP + +SS
Sbjct: 37 QIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIPSPFAN-LSSLRT 213
Query: 105 LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLK 164
L+L+ N+ +G IP S LQVL+L N+ +G +P L T++ NL L+L N L+
Sbjct: 214 LNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLS---NLVTLDLSSNLLE 384
Query: 165 GTIPDMFPASCF-LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNIS 223
G+I + F L L L L + A LE + L I FP +LK S
Sbjct: 385 GSIKESNFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKRQS 564
Query: 224 TLRVLIL 230
+++VL +
Sbjct: 565 SVKVLTM 585
Score = 75.9 bits (185), Expect = 2e-14
Identities = 92/330 (27%), Positives = 143/330 (42%), Gaps = 58/330 (17%)
Frame = +1
Query: 10 LDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLT-DLEGPLQKLNNVSSLHYLDLHNNQLQ 67
LDL NQ+ G LP+ L +L +L L++S+N T + P N+SSL L+L +N+L
Sbjct: 70 LDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIPSPFA---NLSSLRTLNLAHNRLN 240
Query: 68 GPIP---IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDS-LCNA 123
G IP F N+ ++ N + +P +G +S+ + L LS N G+I +S
Sbjct: 241 GTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGT-LSNLVTLDLSSNLLEGSIKESNFVKL 417
Query: 124 TGLQVLDLSINNF-----SGTIPSC---------------------------LMTMAKPE 151
L+ L LS N SG P ++TM+K
Sbjct: 418 FTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKRQSSVKVLTMSKAG 597
Query: 152 --------------NLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQ 197
+E L+L +N L+G + ++F S S +NL N G +P A
Sbjct: 598 IADLVPSWFWIWTLQIEFLDLSNNLLRGDLSNIFLNS---SVINLSSNLFKGRLPSVSA- 765
Query: 198 CSTLEVLDLGKNHITGGFPCFL----KNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQ 253
+EVL++ N I+G FL + L VL NN G LG V+ W+ L
Sbjct: 766 --NVEVLNVANNSISGTISPFLCGKPNATNKLSVLDFSNNVLSGDLGHCWVH---WQALV 930
Query: 254 IMDIAFNNFSGTLKGK--YFKNWETMMHDE 281
+++ NN SG + Y E+++ D+
Sbjct: 931 HVNLGSNNMSGEIPNSMGYLSQLESLLLDD 1020
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 97.1 bits (240), Expect = 9e-21
Identities = 81/288 (28%), Positives = 137/288 (47%), Gaps = 28/288 (9%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
L+ L L+ N+ G +P L L+ L YLN+S+N+ + +L + SL LDL+NN
Sbjct: 294 LSNLSLAANKFSGPIPPSLSALSGLRYLNLSNNVFNETFP--SELWRLQSLEVLDLYNNN 467
Query: 66 LQGPIPIFPV---NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
+ G +P+ N+ ++ N FS IP + G + +L++S N+ G IP + N
Sbjct: 468 MTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQ-YLAVSGNELDGTIPPEIGN 644
Query: 123 ATGLQVLDLS-INNFSGTIPS---------------CLMT------MAKPENLEVLNLRD 160
T L+ L + N ++G IP C ++ + K + L+ L L+
Sbjct: 645 LTSLRELYIGYYNTYTGGIPPEIGNLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQV 824
Query: 161 NNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLK 220
N L G++ L +++L N L G IP S + + +L+L +N + G P F+
Sbjct: 825 NALSGSLTPELGNLKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIG 1004
Query: 221 NISTLRVLILRNNRFQGSL--GCGQVNDEPWKILQIMDIAFNNFSGTL 266
+ L V+ L N GS+ G G+ L ++D++ N +GTL
Sbjct: 1005ELPALEVVQLWENNLTGSIPEGLGKNGR-----LNLVDLSSNKLTGTL 1133
Score = 96.7 bits (239), Expect = 1e-20
Identities = 73/240 (30%), Positives = 123/240 (50%), Gaps = 6/240 (2%)
Frame = +3
Query: 6 TLTYLDLSKNQIHGVLPNWLWEL-NLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNN 64
+L LDL N + GVLP + ++ NL +L++ N + P + L YL + N
Sbjct: 435 SLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPP--EYGRWQRLQYLAVSGN 608
Query: 65 QLQGPIP-----IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDS 119
+L G IP + + +Y+ Y N ++ IP +IG+ +S + L ++ G IP +
Sbjct: 609 ELDGTIPPEIGNLTSLRELYIGYY-NTYTGGIPPEIGN-LSELVRLDVAYCALSGEIPAA 782
Query: 120 LCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLST 179
L L L L +N SG++ L + ++L+ ++L +N L G IP F ++
Sbjct: 783 LGKLQKLDTLFLQVNALSGSLTPELGNL---KSLKSMDLSNNMLSGEIPASFGELKNITL 953
Query: 180 LNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
LNL N+LHG IP+ + + LEV+ L +N++TG P L L ++ L +N+ G+L
Sbjct: 954 LNLFRNKLHGAIPEFIGELPALEVVQLWENNLTGSIPEGLGKNGRLNLVDLSSNKLTGTL 1133
Score = 95.9 bits (237), Expect = 2e-20
Identities = 81/274 (29%), Positives = 123/274 (44%), Gaps = 9/274 (3%)
Frame = +3
Query: 3 NHSTLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYLDL 61
N S L LD++ + G +P L +L L L + N L+ P +L N+ SL +DL
Sbjct: 717 NLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTP--ELGNLKSLKSMDL 890
Query: 62 HNNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
NN L G IP G+ + + L+L NK HG IP+ +
Sbjct: 891 SNNMLSGEIPA---------------------SFGE-LKNITLLNLFRNKLHGAIPEFIG 1004
Query: 122 NATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLN 181
L+V+ L NN +G+IP L K L +++L N L GT+P + L TL
Sbjct: 1005ELPALEVVQLWENNLTGSIPEGL---GKNGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLI 1175
Query: 182 LRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQG---- 237
GN L GPIP+SL C +L + +G+N + G P L + L + L++N G
Sbjct: 1176TLGNFLFGPIPESLGTCESLTRIRMGENFLNGSIPKGLFGLPKLTQVELQDNYLSGEFPE 1355
Query: 238 ----SLGCGQVNDEPWKILQIMDIAFNNFSGTLK 267
++ GQ+ ++ + + NFS K
Sbjct: 1356VGSVAVNLGQITLSNNQLSGALSPSIGNFSSVQK 1457
Score = 83.6 bits (205), Expect = 1e-16
Identities = 75/288 (26%), Positives = 128/288 (44%), Gaps = 26/288 (9%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQK-LNNVSSLHYLDLHNN 64
+T L+L +N++HG +P ++ EL L + + N LT G + + L L+ +DL +N
Sbjct: 945 ITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLT---GSIPEGLGKNGRLNLVDLSSN 1115
Query: 65 QLQGPIPIFPV--NVVYVDYSRNRFS-SVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLC 121
+L G +P + N + + F IP+ +G S + + +N +G+IP L
Sbjct: 1116 KLTGTLPPYLCSGNTLQTLITLGNFLFGPIPESLGT-CESLTRIRMGENFLNGSIPKGLF 1292
Query: 122 NATGLQVLDLSINNFSGTIPSC--------LMTMAKPE-------------NLEVLNLRD 160
L ++L N SG P +T++ + +++ L L
Sbjct: 1293 GLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGALSPSIGNFSSVQKLLLDG 1472
Query: 161 NNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLK 220
N G IP LS ++ GN+ GPI ++QC L LDL +N ++G P +
Sbjct: 1473 NMFTGRIPTQIGRLQQLSKIDFSGNKFSGPIAPEISQCKLLTFLDLSRNELSGDIPNEIT 1652
Query: 221 NISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKG 268
+ L L L N GS+ + + L +D ++NN SG + G
Sbjct: 1653 GMRILNYLNLSKNHLVGSI---PSSISSMQSLTSVDFSYNNLSGLVPG 1787
Score = 69.3 bits (168), Expect = 2e-12
Identities = 59/188 (31%), Positives = 88/188 (46%), Gaps = 4/188 (2%)
Frame = +3
Query: 7 LTYLDLSKNQIHGVLPN-WLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQ 65
LT ++L N + G P +NL + +S+N L+ P + N SS+ L L N
Sbjct: 1305 LTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGALSP--SIGNFSSVQKLLLDGNM 1478
Query: 66 LQGPIPIFPVNVVY---VDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
G IP + +D+S N+FS I +I FL LS N+ G+IP+ +
Sbjct: 1479 FTGRIPTQIGRLQQLSKIDFSGNKFSGPIAPEISQ-CKLLTFLDLSRNELSGDIPNEITG 1655
Query: 123 ATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNL 182
L L+LS N+ G+IPS + +M ++L ++ NNL G +P S F T L
Sbjct: 1656 MRILNYLNLSKNHLVGSIPSSISSM---QSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFL 1826
Query: 183 RGNQLHGP 190
L GP
Sbjct: 1827 GNPDLCGP 1850
Score = 60.5 bits (145), Expect = 9e-10
Identities = 45/150 (30%), Positives = 72/150 (48%), Gaps = 3/150 (2%)
Frame = +3
Query: 133 INNFSGTIPSCL---MTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHG 189
+++++ +IP C +T ++ LNL +L GT+ FLS L+L N+ G
Sbjct: 153 LSSWNASIPYCSWLGVTCDNRRHVTALNLTGLDLSGTLSADVAHLPFLSNLSLAANKFSG 332
Query: 190 PIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPW 249
PIP SL+ S L L+L N FP L + +L VL L NN G L
Sbjct: 333 PIPPSLSALSGLRYLNLSNNVFNETFPSELWRLQSLEVLDLYNNNMTGVLPLAVAQ---M 503
Query: 250 KILQIMDIAFNNFSGTLKGKYFKNWETMMH 279
+ L+ + + N FSG + +Y + W+ + +
Sbjct: 504 QNLRHLHLGGNFFSGQIPPEYGR-WQRLQY 590
>TC205792 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (93%)
Length = 1540
Score = 95.5 bits (236), Expect = 3e-20
Identities = 70/198 (35%), Positives = 103/198 (51%), Gaps = 8/198 (4%)
Frame = +1
Query: 50 LNNVSSLHYLDLHNNQLQGPIP-----IFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIF 104
+ ++ L LDLHNN+L GPIP + + ++ + + N+ IP +IG+ + S
Sbjct: 556 VTSLLDLTRLDLHNNKLTGPIPPQIGRLKRLKILNLRW--NKLQDAIPPEIGE-LKSLTH 726
Query: 105 LSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLK 164
L LS N F G IP L N L+ L L N +G IP L T+ +NL L+ +N+L
Sbjct: 727 LYLSFNNFKGEIPKELANLPDLRYLYLHENRLAGRIPPELGTL---QNLRHLDAGNNHLV 897
Query: 165 GTIPDMFP-ASCF--LSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKN 221
GTI ++ CF L L L N G IP LA ++LE+L L N ++G P + +
Sbjct: 898 GTIRELIRIEGCFPALRNLYLNNNYFTGGIPAQLANLTSLEILYLSYNKMSGVIPSTVAH 1077
Query: 222 ISTLRVLILRNNRFQGSL 239
I L L L +N+F G +
Sbjct: 1078IPKLTYLYLDHNQFSGRI 1131
Score = 59.7 bits (143), Expect = 2e-09
Identities = 48/153 (31%), Positives = 67/153 (43%)
Frame = +1
Query: 87 FSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMT 146
+S V +GDY L + G P ++ + L LDL N +G IP
Sbjct: 460 WSGVTCSTVGDYRV-VTELEVYAVSIVGPFPTAVTSLLDLTRLDLHNNKLTGPIPP---Q 627
Query: 147 MAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDL 206
+ + + L++LNLR N L+ IP L+ L L N G IPK LA L L L
Sbjct: 628 IGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNNFKGEIPKELANLPDLRYLYL 807
Query: 207 GKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
+N + G P L + LR L NN G++
Sbjct: 808 HENRLAGRIPPELGTLQNLRHLDAGNNHLVGTI 906
>TC220443 weakly similar to UP|Q9S711 (Q9S711) ESTs C22657(S0014)
(Transmembrane protein kinase), partial (8%)
Length = 634
Score = 92.8 bits (229), Expect = 2e-19
Identities = 66/191 (34%), Positives = 92/191 (47%), Gaps = 3/191 (1%)
Frame = +3
Query: 52 NVSSLHYLDLHNNQLQGPIPI---FPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLS 108
N S L L HNN+ G IP N+ SRN+F+ V+P+ + +S +S
Sbjct: 3 NCSGLLDLKAHNNEFSGNIPSGLWTSFNLTNFMVSRNKFTGVLPERLSWNISR---FEIS 173
Query: 109 DNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIP 168
N+F G IP + + T L V D S NNF+G+IP L + K L L L N L G +P
Sbjct: 174 YNQFSGGIPSGVSSWTNLVVFDASKNNFNGSIPRQLTALPK---LTTLLLDQNQLTGELP 344
Query: 169 DMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVL 228
+ L LNL NQL+G IP ++ Q L LDL +N +G P ++ L
Sbjct: 345 SDIISWKSLVALNLSQNQLYGQIPHAIGQLPALSQLDLSENEFSGQVPSLPPRLTNLN-- 518
Query: 229 ILRNNRFQGSL 239
L +N G +
Sbjct: 519 -LSSNHLTGRI 548
>BQ081034
Length = 421
Score = 91.7 bits (226), Expect = 4e-19
Identities = 55/137 (40%), Positives = 80/137 (58%)
Frame = +2
Query: 99 MSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNL 158
+SS LSL GN+P+S+ + L++L++S N+F+G IPS L + NL+ + L
Sbjct: 20 LSSLKVLSLVSLGLWGNLPESIAQLSSLEILNISSNHFNGAIPSQLSLL---RNLQSVVL 190
Query: 159 RDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCF 218
DNN G I + + L+ L++R N L G +P SL TL VLDL N ++G P
Sbjct: 191 DDNNFNGEISNWVGSLQGLAVLSMRNNWLSGSLPTSLNALHTLRVLDLSNNQLSGELP-H 367
Query: 219 LKNISTLRVLILRNNRF 235
LKN++ L+VL L NN F
Sbjct: 368 LKNLANLQVLNLENNTF 418
Score = 59.3 bits (142), Expect = 2e-09
Identities = 38/121 (31%), Positives = 58/121 (47%)
Frame = +2
Query: 119 SLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLS 178
+L + L+VL L G +P ++A+ +LE+LN+ N+ G IP L
Sbjct: 8 TLGTLSSLKVLSLVSLGLWGNLPE---SIAQLSSLEILNISSNHFNGAIPSQLSLLRNLQ 178
Query: 179 TLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGS 238
++ L N +G I + L VL + N ++G P L + TLRVL L NN+ G
Sbjct: 179 SVVLDDNNFNGEISNWVGSLQGLAVLSMRNNWLSGSLPTSLNALHTLRVLDLSNNQLSGE 358
Query: 239 L 239
L
Sbjct: 359 L 361
Score = 35.8 bits (81), Expect = 0.024
Identities = 24/62 (38%), Positives = 34/62 (54%), Gaps = 2/62 (3%)
Frame = +2
Query: 12 LSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQ-KLNNVSSLHYLDLHNNQLQGP 69
L N +G + NW+ L L L++ +N L+ G L LN + +L LDL NNQL G
Sbjct: 188 LDDNNFNGEISNWVGSLQGLAVLSMRNNWLS---GSLPTSLNALHTLRVLDLSNNQLSGE 358
Query: 70 IP 71
+P
Sbjct: 359 LP 364
Score = 31.2 bits (69), Expect = 0.59
Identities = 16/33 (48%), Positives = 20/33 (60%)
Frame = +2
Query: 6 TLTYLDLSKNQIHGVLPNWLWELNLHYLNISSN 38
TL LDLS NQ+ G LP+ NL LN+ +N
Sbjct: 314 TLRVLDLSNNQLSGELPHLKNLANLQVLNLENN 412
>TC208419 weakly similar to UP|Q6X1D9 (Q6X1D9) Resistance protein SlVe1
precursor, partial (9%)
Length = 1140
Score = 90.9 bits (224), Expect = 6e-19
Identities = 61/194 (31%), Positives = 84/194 (42%), Gaps = 27/194 (13%)
Frame = +1
Query: 76 NVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINN 135
++V+++ N FS V+P + M I L N+F G IP C+ L LDLS N
Sbjct: 67 SLVFINLGENNFSGVLPTKMPKSMQVMI---LRSNQFAGKIPPETCSLPSLSQLDLSQNK 237
Query: 136 FSGTIPSCLMTMAKPEN---------------------------LEVLNLRDNNLKGTIP 168
SG+IP C+ + + + L+ L+ NNL G IP
Sbjct: 238 LSGSIPPCVYNITRMDGERRASHFQFSLDLFWKGRELQYKDTGLLKNLDFSTNNLSGEIP 417
Query: 169 DMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVL 228
+ L LNL N L G IP + LE LDL NH++G P + N+S L L
Sbjct: 418 PELFSLTELLFLNLSRNNLMGKIPSKIGGMKNLESLDLSNNHLSGEIPAAISNLSFLSYL 597
Query: 229 ILRNNRFQGSLGCG 242
L N F G + G
Sbjct: 598 NLSYNDFTGQIPLG 639
>TC228573
Length = 1628
Score = 90.1 bits (222), Expect = 1e-18
Identities = 71/209 (33%), Positives = 101/209 (47%), Gaps = 5/209 (2%)
Frame = +3
Query: 3 NHSTLTYLDLSKNQIHGVLPNWLWELN-LHYLNISSNLLTDLEGPLQKLNNVSSLHYLDL 61
N + L L L+ N+ G +P+ + +L L L + +N LT Q + + +L YL L
Sbjct: 486 NLTRLDVLSLTGNRFIGPVPSSITKLTQLTQLKLGNNFLTGTVP--QGIAKLVNLTYLSL 659
Query: 62 HNNQLQGPIPIFP---VNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPD 118
NQL+G IP F ++ +++S N+FS IP I +L L N G IPD
Sbjct: 660 EGNQLEGTIPDFFSSFTDLRILNFSYNKFSGNIPNSISSLAPKLTYLELGHNSLSGKIPD 839
Query: 119 SLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLS 178
L L LDLS N FSGT+P+ + K N LNL +N L P+M +
Sbjct: 840 FLGKFKALDTLDLSWNKFSGTVPASFKNLTKIFN---LNLSNNLLVDPFPEMNVKG--IE 1004
Query: 179 TLNLRGNQLH-GPIPKSLAQCSTLEVLDL 206
+L+L N H G IPK +A + L L
Sbjct: 1005SLDLSNNSFHLGSIPKWVASSPIIFSLKL 1091
Score = 88.2 bits (217), Expect = 4e-18
Identities = 70/215 (32%), Positives = 98/215 (45%), Gaps = 28/215 (13%)
Frame = +3
Query: 47 LQKLNNVSSLHYLDLHNNQLQGPIPIFPV---NVVYVDYSRNRFSSVIPQDIGDYMSSAI 103
L KL + L+ ++L N + GP P F N+ ++ N S IP +IG+ ++
Sbjct: 333 LSKLKLLDGLYLINLIN--ISGPFPNFLFQLPNLQFIYLENNNLSGRIPDNIGN-LTRLD 503
Query: 104 FLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNL 163
LSL+ N+F G +P S+ T L L L N +GT+P + AK NL L+L N L
Sbjct: 504 VLSLTGNRFIGPVPSSITKLTQLTQLKLGNNFLTGTVPQGI---AKLVNLTYLSLEGNQL 674
Query: 164 KGTIPDMFPASCFLSTLN-------------------------LRGNQLHGPIPKSLAQC 198
+GTIPD F + L LN L N L G IP L +
Sbjct: 675 EGTIPDFFSSFTDLRILNFSYNKFSGNIPNSISSLAPKLTYLELGHNSLSGKIPDFLGKF 854
Query: 199 STLEVLDLGKNHITGGFPCFLKNISTLRVLILRNN 233
L+ LDL N +G P KN++ + L L NN
Sbjct: 855 KALDTLDLSWNKFSGTVPASFKNLTKIFNLNLSNN 959
Score = 85.5 bits (210), Expect = 3e-17
Identities = 59/173 (34%), Positives = 89/173 (51%), Gaps = 3/173 (1%)
Frame = +3
Query: 104 FLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNL 163
F+ L +N G IPD++ N T L VL L+ N F G +PS ++ K L L L +N L
Sbjct: 432 FIYLENNNLSGRIPDNIGNLTRLDVLSLTGNRFIGPVPS---SITKLTQLTQLKLGNNFL 602
Query: 164 KGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNIS 223
GT+P L+ L+L GNQL G IP + + L +L+ N +G P + +++
Sbjct: 603 TGTVPQGIAKLVNLTYLSLEGNQLEGTIPDFFSSFTDLRILNFSYNKFSGNIPNSISSLA 782
Query: 224 -TLRVLILRNNRFQGSLG--CGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKN 273
L L L +N G + G+ +K L +D+++N FSGT+ FKN
Sbjct: 783 PKLTYLELGHNSLSGKIPDFLGK-----FKALDTLDLSWNKFSGTVPAS-FKN 923
Score = 72.8 bits (177), Expect = 2e-13
Identities = 49/163 (30%), Positives = 83/163 (50%), Gaps = 10/163 (6%)
Frame = +3
Query: 114 GNIPDSLCNAT------GLQVLD----LSINNFSGTIPSCLMTMAKPENLEVLNLRDNNL 163
G P+++ + T L++LD +++ N SG P+ L + NL+ + L +NNL
Sbjct: 288 GQKPETILSGTISPTLSKLKLLDGLYLINLINISGPFPNFLFQLP---NLQFIYLENNNL 458
Query: 164 KGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNIS 223
G IPD L L+L GN+ GP+P S+ + + L L LG N +TG P + +
Sbjct: 459 SGRIPDNIGNLTRLDVLSLTGNRFIGPVPSSITKLTQLTQLKLGNNFLTGTVPQGIAKLV 638
Query: 224 TLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTL 266
L L L N+ +G++ + L+I++ ++N FSG +
Sbjct: 639 NLTYLSLEGNQLEGTI---PDFFSSFTDLRILNFSYNKFSGNI 758
Score = 28.9 bits (63), Expect = 2.9
Identities = 17/40 (42%), Positives = 24/40 (59%)
Frame = +3
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWELNLHYLNISSNLL 40
LR L +LDLS+N + G +PN + + L LN+S N L
Sbjct: 1272 LRFGERLKFLDLSRNWVFGKVPNSV--VGLEKLNVSYNHL 1385
>BG654265 weakly similar to GP|3894385|gb|A Hcr2-0A {Lycopersicon
esculentum}, partial (6%)
Length = 416
Score = 89.7 bits (221), Expect = 1e-18
Identities = 49/114 (42%), Positives = 67/114 (57%), Gaps = 1/114 (0%)
Frame = +3
Query: 165 GTIPDMFPASCFLSTLNLRGNQL-HGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNIS 223
GT+P F C L L+L GNQL G +P+SL+ CS L VLDLG N I FP +L+ +
Sbjct: 3 GTLPSTFAQDCGLKILDLNGNQLLEGFLPESLSNCSLLRVLDLGNNQIKDVFPHWLQTLL 182
Query: 224 TLRVLILRNNRFQGSLGCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKNWETM 277
L+VL+LR N+ G + G + + L I D++ NNFSG + Y K +E M
Sbjct: 183 DLKVLVLRANKLYGPI-AGSKTKDAFPSLVIFDVSSNNFSGPIPKAYIKKFEAM 341
Score = 47.8 bits (112), Expect = 6e-06
Identities = 36/96 (37%), Positives = 49/96 (50%), Gaps = 6/96 (6%)
Frame = +3
Query: 105 LSLSDNKF-HGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTMAKPENLEVLNLRDNNL 163
L L+ N+ G +P+SL N + L+VLDL N P L T+ +L+VL LR N L
Sbjct: 48 LDLNGNQLLEGFLPESLSNCSLLRVLDLGNNQIKDVFPHWLQTLL---DLKVLVLRANKL 218
Query: 164 KGTI-----PDMFPASCFLSTLNLRGNQLHGPIPKS 194
G I D FP+ L ++ N GPIPK+
Sbjct: 219 YGPIAGSKTKDAFPS---LVIFDVSSNNFSGPIPKA 317
Score = 39.7 bits (91), Expect = 0.002
Identities = 27/72 (37%), Positives = 35/72 (48%), Gaps = 1/72 (1%)
Frame = +3
Query: 1 LRNHSTLTYLDLSKNQIHGVLPNWLWE-LNLHYLNISSNLLTDLEGPLQKLNNVSSLHYL 59
L N S L LDL NQI V P+WL L+L L + +N L + + SL
Sbjct: 96 LSNCSLLRVLDLGNNQIKDVFPHWLQTLLDLKVLVLRANKLYGPIAGSKTKDAFPSLVIF 275
Query: 60 DLHNNQLQGPIP 71
D+ +N GPIP
Sbjct: 276 DVSSNNFSGPIP 311
Score = 37.4 bits (85), Expect = 0.008
Identities = 40/127 (31%), Positives = 56/127 (43%), Gaps = 4/127 (3%)
Frame = +3
Query: 19 GVLPN-WLWELNLHYLNISSNLLTDLEGPL-QKLNNVSSLHYLDLHNNQLQGPIPIFPVN 76
G LP+ + + L L+++ N L LEG L + L+N S L LDL NNQ++
Sbjct: 3 GTLPSTFAQDCGLKILDLNGNQL--LEGFLPESLSNCSLLRVLDLGNNQIK--------- 149
Query: 77 VVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNAT--GLQVLDLSIN 134
V P + + + L L NK +G I S L + D+S N
Sbjct: 150 ------------DVFPHWLQTLLDLKV-LVLRANKLYGPIAGSKTKDAFPSLVIFDVSSN 290
Query: 135 NFSGTIP 141
NFSG IP
Sbjct: 291 NFSGPIP 311
>TC212367 weakly similar to UP|Q9ZTJ6 (Q9ZTJ6) Hcr2-5D, partial (11%)
Length = 787
Score = 84.7 bits (208), Expect = 5e-17
Identities = 63/180 (35%), Positives = 90/180 (50%), Gaps = 5/180 (2%)
Frame = +2
Query: 19 GVLPNWLWELNLHYLNISSNLLTDLEGPLQKLNNVSSLHYLDLHNNQLQGPIP-IFPV-- 75
G +P+W+ L+ YL S+ D P + L N+ L LDLH+N+L G I +F
Sbjct: 5 GSIPSWIGSLSQLYLLXXSSNSLDSHIP-ESLTNLPDLGVLDLHSNKLTGSIAGVFDTEQ 181
Query: 76 -NVVYVDYSRNRFSSVIPQ-DIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSI 133
+ Y+D S N FSS + +G+ ++ +L+LS N G +P SL + LDLS
Sbjct: 182 GTLTYIDLSDNNFSSGVEAIGVGEQLNIQ-YLNLSHNLLKGTLPSSLGKLNSIHSLDLSF 358
Query: 134 NNFSGTIPSCLMTMAKPENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPK 193
N + +P L AK LE L L+ N+ G IP F L L+L N L G IP+
Sbjct: 359 NELASNLPEML---AKLTLLERLKLQGNHFSGKIPSGFLKLKKLKELDLSDNVLEGEIPE 529
Score = 72.4 bits (176), Expect = 2e-13
Identities = 53/170 (31%), Positives = 77/170 (45%), Gaps = 1/170 (0%)
Frame = +2
Query: 88 SSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIPSCLMTM 147
S IP IG +S L S N +IP+SL N L VLDL N +G+I T
Sbjct: 2 SGSIPSWIGS-LSQLYLLXXSSNSLDSHIPESLTNLPDLGVLDLHSNKLTGSIAGVFDT- 175
Query: 148 AKPENLEVLNLRDNNLKGTIPDMFPASCF-LSTLNLRGNQLHGPIPKSLAQCSTLEVLDL 206
+ L ++L DNN + + + LNL N L G +P SL + +++ LDL
Sbjct: 176 -EQGTLTYIDLSDNNFSSGVEAIGVGEQLNIQYLNLSHNLLKGTLPSSLGKLNSIHSLDL 352
Query: 207 GKNHITGGFPCFLKNISTLRVLILRNNRFQGSLGCGQVNDEPWKILQIMD 256
N + P L ++ L L L+ N F G + G + + K L + D
Sbjct: 353 SFNELASNLPEMLAKLTLLERLKLQGNHFSGKIPSGFLKLKKLKELDLSD 502
Score = 54.3 bits (129), Expect = 7e-08
Identities = 49/139 (35%), Positives = 60/139 (42%), Gaps = 3/139 (2%)
Frame = +2
Query: 6 TLTYLDLSKNQI-HGVLPNWLWE-LNLHYLNISSNLLT-DLEGPLQKLNNVSSLHYLDLH 62
TLTY+DLS N GV + E LN+ YLN+S NLL L L KLN S+H LDL
Sbjct: 185 TLTYIDLSDNNFSSGVEAIGVGEQLNIQYLNLSHNLLKGTLPSSLGKLN---SIHSLDLS 355
Query: 63 NNQLQGPIPIFPVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLSLSDNKFHGNIPDSLCN 122
N+L +P + ++ L L N F G IP
Sbjct: 356 FNELASNLPEMLAKLTLLER----------------------LKLQGNHFSGKIPSGFLK 469
Query: 123 ATGLQVLDLSINNFSGTIP 141
L+ LDLS N G IP
Sbjct: 470 LKKLKELDLSDNVLEGEIP 526
>TC224271 weakly similar to UP|Q84XU7 (Q84XU7) Receptor-like protein kinase,
partial (19%)
Length = 783
Score = 83.6 bits (205), Expect = 1e-16
Identities = 62/218 (28%), Positives = 95/218 (43%), Gaps = 28/218 (12%)
Frame = +1
Query: 50 LNNVSSLHYLDLHNNQLQGPIPIF---PVNVVYVDYSRNRFSSVIPQDIGDYMSSAIFLS 106
L ++ YL L N +P + + +D S N F+ +P IG+ + L+
Sbjct: 7 LKELTLCGYLSLRGNAFSREVPEWIGEMRGLETLDLSNNGFTGQVPSSIGN-LQLLKMLN 183
Query: 107 LSDNKFHGNIPDSLCNATGLQVLDLSINNFSGTIP-----------------------SC 143
S N G++P+S+ N T L VLD+S N+ SG +P S
Sbjct: 184 FSGNGLTGSLPESIVNCTKLSVLDVSRNSMSGWLPLWVFKSDLDKGLMSENVQSGSKKSP 363
Query: 144 LMTMAKP--ENLEVLNLRDNNLKGTIPDMFPASCFLSTLNLRGNQLHGPIPKSLAQCSTL 201
L +A+ ++L+VL+L N G I L LNL N L GPIP ++ + T
Sbjct: 364 LFALAEVAFQSLQVLDLSHNAFSGEITSAVGGLSSLQVLNLANNSLGGPIPAAIGELKTC 543
Query: 202 EVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
LDL N + G P + +L+ L+L N G +
Sbjct: 544 SSLDLSYNKLNGSIPWEIGRAVSLKELVLEKNFLNGKI 657
Score = 50.4 bits (119), Expect = 9e-07
Identities = 32/94 (34%), Positives = 47/94 (49%)
Frame = +1
Query: 180 LNLRGNQLHGPIPKSLAQCSTLEVLDLGKNHITGGFPCFLKNISTLRVLILRNNRFQGSL 239
L+LRGN +P+ + + LE LDL N TG P + N+ L++L N GSL
Sbjct: 34 LSLRGNAFSREVPEWIGEMRGLETLDLSNNGFTGQVPSSIGNLQLLKMLNFSGNGLTGSL 213
Query: 240 GCGQVNDEPWKILQIMDIAFNNFSGTLKGKYFKN 273
VN L ++D++ N+ SG L FK+
Sbjct: 214 PESIVNCTK---LSVLDVSRNSMSGWLPLWVFKS 306
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,087,197
Number of Sequences: 63676
Number of extensions: 226035
Number of successful extensions: 1971
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 1290
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1635
length of query: 299
length of database: 12,639,632
effective HSP length: 97
effective length of query: 202
effective length of database: 6,463,060
effective search space: 1305538120
effective search space used: 1305538120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0490.10


