

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0487.5
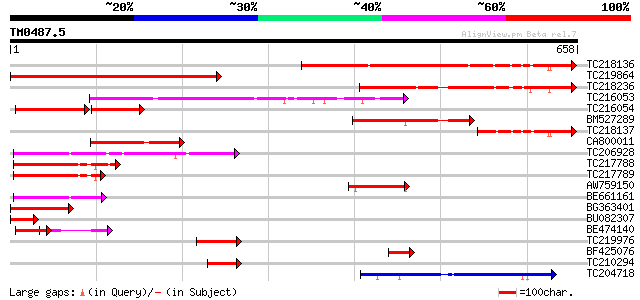
(658 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218136 484 e-137
TC219864 424 e-119
TC218236 317 1e-86
TC216053 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (33%) 219 3e-57
TC216054 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (36%) 117 1e-47
BM527289 183 2e-46
TC218137 143 3e-34
CA800011 115 5e-26
TC206928 similar to UP|Q9LNC6 (Q9LNC6) F9P14.7 protein (At1g0621... 102 7e-22
TC217788 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like... 99 6e-21
TC217789 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like... 87 2e-17
AW759150 similar to GP|21430672|gb| RE65163p {Drosophila melanog... 79 5e-15
BE661161 72 8e-13
BG363401 71 1e-12
BU082307 65 1e-10
BE474140 59 7e-09
TC219976 similar to UP|OAZ_HUMAN (P54368) Ornithine decarboxylas... 54 2e-07
BF425076 similar to GP|28381653|gb| CG1693-PA {Drosophila melano... 52 8e-07
TC210294 45 1e-04
TC204718 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17... 45 1e-04
>TC218136
Length = 1097
Score = 484 bits (1246), Expect = e-137
Identities = 246/329 (74%), Positives = 278/329 (83%), Gaps = 10/329 (3%)
Frame = +1
Query: 339 IDPKLDLLSGDDYNSPKADNSLALVPVGQQQPASPMSQQNALVLFDMFSNGSNAP-SVNN 397
+DPK+DLLSGDDYNSPKA+ SLALVP+G+QQPASPMSQQNALVLFDMFSNGSNAP SVN
Sbjct: 1 VDPKVDLLSGDDYNSPKAETSLALVPLGEQQPASPMSQQNALVLFDMFSNGSNAPISVNT 180
Query: 398 QPTNVAVPNSPLSPQFQQQQTFISQGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNGQAA 457
QP N+ SPL+PQFQQQ TFISQGV+YPNGS+P+VGSP+YEQS +AQSTGP+WNGQ A
Sbjct: 181 QPINIVGQTSPLAPQFQQQ-TFISQGVFYPNGSVPNVGSPRYEQSPFAQSTGPSWNGQVA 357
Query: 458 QQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQ 517
QQQQPPSPV G SGGSFPPPPWEAQ +N SPVAG+QYPQPLQV+QM++T +Q PQ
Sbjct: 358 QQQQPPSPVYGTASGGSFPPPPWEAQPTDNNSPVAGSQYPQPLQVSQMIMTSIQSGTHPQ 537
Query: 518 GPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQN 577
GPQAMG+DQAVGMYM QP+A HMS +NN VQSNQ G++ Q+ QG AA PYMGMV HQM N
Sbjct: 538 GPQAMGHDQAVGMYM-QPHAGHMSTINNHVQSNQLGLYRQHIQG-AAGPYMGMVSHQMHN 711
Query: 578 GPVAHPMYAQQMYGNQYMGGYGYGQQPQQGVQYVEQQMHGLSMSDD---RNS------SY 628
PVA MY QQM+GNQ+ GGYGYGQQP GVQY+EQQM+GLS+ DD +NS SY
Sbjct: 712 SPVA-SMYPQQMHGNQF-GGYGYGQQP--GVQYLEQQMYGLSVRDDGAPKNSYQVSSPSY 879
Query: 629 VPPKKPSKPEDKLFGDLVDMAKVKPKPTP 657
VPP KPSKPEDKLFGDLV+MAKVKPKPTP
Sbjct: 880 VPPGKPSKPEDKLFGDLVNMAKVKPKPTP 966
>TC219864
Length = 932
Score = 424 bits (1091), Expect = e-119
Identities = 208/245 (84%), Positives = 230/245 (92%)
Frame = +3
Query: 1 MVNPLVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALT 60
MVN +VERATSDMLIGPDWA+NIEICD+LN DP Q+KDVVKG+KKRIGS++SKVQLLALT
Sbjct: 198 MVNSMVERATSDMLIGPDWAMNIEICDMLNHDPGQAKDVVKGIKKRIGSKNSKVQLLALT 377
Query: 61 LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRVREKIMILIDTWQEAFGGPRARY 120
LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPD V+EKI++L+DTWQEAFGGPRARY
Sbjct: 378 LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDFHVKEKILVLVDTWQEAFGGPRARY 557
Query: 121 PQYYAAYQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQNIRDSDAQPHTAESSAESE 180
PQYYAAYQELL +GAVFPQRSEQSAPVFTP QTQPL SYPQNI D++ + A+SSAESE
Sbjct: 558 PQYYAAYQELLRAGAVFPQRSEQSAPVFTPPQTQPLASYPQNIPDTNVDFYAAQSSAESE 737
Query: 181 FPTLSLSEIQNARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLVNST 240
FPTL+L+EIQNARGIMDVLAEML+A++P NKEG+RQEVI DLVEQCRTYKQRVVHLVNST
Sbjct: 738 FPTLNLTEIQNARGIMDVLAEMLNALDPSNKEGIRQEVIADLVEQCRTYKQRVVHLVNST 917
Query: 241 SDESL 245
SDESL
Sbjct: 918 SDESL 932
>TC218236
Length = 1198
Score = 317 bits (811), Expect = 1e-86
Identities = 172/269 (63%), Positives = 192/269 (70%), Gaps = 18/269 (6%)
Frame = +2
Query: 407 SPLSPQFQQQQTFISQGVYYPNGSMPSVGSPQ-YEQSRYAQSTGPAWNGQAAQQQQ--PP 463
SP +PQFQQQQT SQG +YPNG+ P+ GSPQ YEQS Y Q+TGPAWNGQ AQQQQ PP
Sbjct: 11 SPYAPQFQQQQTVTSQGGFYPNGNAPNAGSPQQYEQSLYTQNTGPAWNGQVAQQQQQQPP 190
Query: 464 SPVNGAQSGGSFPPPPWEAQ-AAENGSPVAGNQYPQPLQVTQMVVTHVQGAPLPQGPQAM 522
SPV G QSG S PPPPWEAQ AA+NGSP+AG QYP H+Q + P GPQ M
Sbjct: 191 SPVYGTQSG-SLPPPPWEAQPAADNGSPLAGAQYP-----------HMQNSGHPMGPQTM 334
Query: 523 GYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGPVAH 582
G DQ VGMYMQ SHMS MNN V SNQ G+ PQ+ QGV A PYMGM PHQMQ GPV
Sbjct: 335 GNDQGVGMYMQPNANSHMSGMNNHVGSNQMGLQPQHMQGV-AGPYMGMAPHQMQGGPV-- 505
Query: 583 PMYAQQMYGNQYMGGYGYGQQ-----PQQGVQYVEQQMHGLSMSDDRN---------SSY 628
MY QQMYGNQ+M GYGY Q+ QQGV Y+E+QM+G+S+ DD + +SY
Sbjct: 506 -MYPQQMYGNQFM-GYGYDQRQGVHYQQQGVPYIERQMYGMSVRDDSSLRNPYQASTTSY 679
Query: 629 VPPKKPSKPEDKLFGDLVDMAKVKPKPTP 657
P KPSKPEDKLFGDLVDMAKVKPKPTP
Sbjct: 680 APSGKPSKPEDKLFGDLVDMAKVKPKPTP 766
>TC216053 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (33%)
Length = 1485
Score = 219 bits (558), Expect = 3e-57
Identities = 156/415 (37%), Positives = 208/415 (49%), Gaps = 44/415 (10%)
Frame = +3
Query: 93 KPDPRVREKIMILIDTWQEAFGGPRARYPQYYAAYQELLHSGAVFPQRSEQSAPVFTPLQ 152
+PD VREKI+ILIDTWQEAFGGP YPQYYAAY EL +G FP R E S P FTP Q
Sbjct: 6 EPDLNVREKILILIDTWQEAFGGPTGVYPQYYAAYNELKSAGVEFPPRDENSVPFFTPAQ 185
Query: 153 TQPLTSYPQNIRDSDAQPHTAESSAESEFPTLSLSEIQNARGIMDVLAEMLSAIEPGNKE 212
TQP+ D+ T ++S +S+ LSL EIQNA+G+ DVL EMLSA+ P ++E
Sbjct: 186 TQPIIHSAAEYDDA-----TIQASLQSDASDLSLLEIQNAQGLADVLMEMLSALSPKDRE 350
Query: 213 GLRQEVIVDLVEQCRTYKQRVVHLVNSTSDESLLCQGLALNDDLQRVLAKHESIASGTPG 272
G+++EVIVDLV+QCR+Y++RV+ LVN+T+DE LL QGLALND LQRVL +H+ I GT
Sbjct: 351 GVKEEVIVDLVDQCRSYQKRVMLLVNNTTDEQLLGQGLALNDSLQRVLCRHDDIVKGTAD 530
Query: 273 QNHTEKLNPAPPGALIDVGGPLVDTGDTSKQSDERSPSGAEGGSQ----------TLNQL 322
E P L++V ++ D Q RS + +Q +N
Sbjct: 531 SGAREAETSVLP--LVNVNHEDDESEDDFAQLAHRSSRDTQAQNQKPAYDKAEPGRINP- 701
Query: 323 LLPAPPTSNGSAPPIKIDPKLDLLSGDDY---NSPKADNSLALV--------PVGQQQPA 371
L+P PP S P +D LSGD Y SP+ + P P+
Sbjct: 702 LIPPPPAS--KKPVYSGTGMVDYLSGDTYKTEGSPENSEPTSFTAPLHSSPNPTSSTIPS 875
Query: 372 SPMSQQNALVLFDMFSNGSNAPSVNNQPTNVAVPNS-----------------------P 408
S +A+ + +++P +++P P+S P
Sbjct: 876 LSSSHPHAV-------STTSSPIFSSEPVYDEQPSSEDKSSECLPAAPWDAQSPGIIPPP 1034
Query: 409 LSPQFQQQQTFISQGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNGQAAQQQQPP 463
S Q+QQ F QGV S S GS S Q+ + N +QQ P
Sbjct: 1035PSKYNQRQQFFEQQGV-----SHSSSGSSSSYDSLVGQTQNLSLNSSTPTKQQKP 1184
Score = 34.3 bits (77), Expect = 0.18
Identities = 49/211 (23%), Positives = 79/211 (37%), Gaps = 9/211 (4%)
Frame = +3
Query: 451 AWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPV----AGNQY-----PQPLQ 501
A N + A + P +N PPPP + +G+ + +G+ Y P+ +
Sbjct: 648 AQNQKPAYDKAEPGRINPL-----IPPPPASKKPVYSGTGMVDYLSGDTYKTEGSPENSE 812
Query: 502 VTQMVVTHVQGAPLPQGPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQFGMHPQYAQG 561
T APL P + P+A +S ++ + S++ Q +
Sbjct: 813 PTSFT------APLHSSPNPTS-STIPSLSSSHPHA--VSTTSSPIFSSEPVYDEQPSSE 965
Query: 562 VAASPYMGMVPHQMQNGPVAHPMYAQQMYGNQYMGGYGYGQQPQQGVQYVEQQMHGLSMS 621
+S + P Q+ + P ++ Q+ G G + G + +
Sbjct: 966 DKSSECLPAAPWDAQSPGIIPPPPSKYNQRQQFFEQQGVSHS-SSGSSSSYDSLVGQTQN 1142
Query: 622 DDRNSSYVPPKKPSKPEDKLFGDLVDMAKVK 652
NSS P K KPED LF DLVD AK K
Sbjct: 1143LSLNSS--TPTKQQKPEDALFKDLVDFAKSK 1229
>TC216054 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (36%)
Length = 713
Score = 117 bits (294), Expect(2) = 1e-47
Identities = 55/86 (63%), Positives = 69/86 (79%)
Frame = +1
Query: 7 ERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLETII 66
ERAT DML GPDWA+NIE+ DI+N DP Q+KD +K LKKR+ S++ ++QLLAL LET+
Sbjct: 223 ERATRDMLSGPDWAINIEVGDIINMDPRQAKDAIKILKKRLSSKNPQIQLLALFALETLS 402
Query: 67 KNCGDIVHMHVAERDVLHEMVKIVKK 92
KNCGD V + E+D+LHEMVKIVKK
Sbjct: 403 KNCGDSVFQQIIEQDILHEMVKIVKK 480
Score = 91.3 bits (225), Expect(2) = 1e-47
Identities = 43/62 (69%), Positives = 47/62 (75%)
Frame = +3
Query: 95 DPRVREKIMILIDTWQEAFGGPRARYPQYYAAYQELLHSGAVFPQRSEQSAPVFTPLQTQ 154
D RVREKI+ILIDTWQEAFGGP +YPQY AAY EL +G FP R E SAP FTP QT
Sbjct: 480 DLRVREKILILIDTWQEAFGGPSGKYPQYLAAYNELKSAGVEFPPREENSAPFFTPPQTL 659
Query: 155 PL 156
P+
Sbjct: 660 PV 665
>BM527289
Length = 422
Score = 183 bits (465), Expect = 2e-46
Identities = 93/147 (63%), Positives = 101/147 (68%), Gaps = 5/147 (3%)
Frame = +2
Query: 398 QPTNVAVPNSPLSPQFQQQQTFI-SQGVYYPNGSMPSVGSPQYEQSRYAQSTGPAWNGQA 456
QPTNVA SP PQFQQQQT + SQG +YPNG+ P+ GSPQYEQS Y QSTGPAWNGQ
Sbjct: 14 QPTNVAGQTSPYGPQFQQQQTVVTSQGGFYPNGNAPNAGSPQYEQSLYTQSTGPAWNGQV 193
Query: 457 A----QQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVTHVQG 512
A QQQQPPSPV G+Q GS PPPPWEAQ A+NG P+AG QYP H+Q
Sbjct: 194 AQQQQQQQQPPSPVYGSQGSGSLPPPPWEAQPADNGGPLAGTQYP-----------HMQN 340
Query: 513 APLPQGPQAMGYDQAVGMYMQQPNASH 539
A P GPQ MG DQ VGMYMQ SH
Sbjct: 341 AGHPMGPQTMGNDQGVGMYMQSNANSH 421
>TC218137
Length = 434
Score = 143 bits (360), Expect = 3e-34
Identities = 82/124 (66%), Positives = 94/124 (75%), Gaps = 9/124 (7%)
Frame = +2
Query: 543 MNNQVQSNQFGMHPQYAQGVAASPYMGMVPHQMQNGPVAHPMYAQQMYGNQYMGGYGYGQ 602
+NN VQSNQ G++PQ QGVA S YM MV HQM N PVA MY QQMYGNQ+ GGYGYGQ
Sbjct: 5 INNHVQSNQLGLYPQNIQGVAGS-YMDMVSHQMHNSPVAS-MYPQQMYGNQF-GGYGYGQ 175
Query: 603 QPQQGVQYVEQQMHGLSMSDD---RNS------SYVPPKKPSKPEDKLFGDLVDMAKVKP 653
QP+ VQ+VEQQM+GLS++ D RNS SYVPP KPSK ED+L+ DLV+MAKVKP
Sbjct: 176 QPR--VQHVEQQMYGLSVTHDGALRNSNQVTSTSYVPPGKPSKTEDELYRDLVNMAKVKP 349
Query: 654 KPTP 657
K TP
Sbjct: 350 KFTP 361
>CA800011
Length = 436
Score = 115 bits (289), Expect = 5e-26
Identities = 60/110 (54%), Positives = 75/110 (67%)
Frame = +3
Query: 94 PDPRVREKIMILIDTWQEAFGGPRARYPQYYAAYQELLHSGAVFPQRSEQSAPVFTPLQT 153
PD RVREKI+ILIDTWQEAFGGP +YPQY AAY EL +G FP R E SAP FTP QT
Sbjct: 120 PDLRVREKILILIDTWQEAFGGPSGKYPQYLAAYNELKSAGVEFPPREENSAPFFTPPQT 299
Query: 154 QPLTSYPQNIRDSDAQPHTAESSAESEFPTLSLSEIQNARGIMDVLAEML 203
P+ ++ ++ + ++S S+ LSL EIQNA+G+ DVL EM+
Sbjct: 300 LPV-----HLAAAEYDDASIQASLHSDASGLSLPEIQNAQGLADVLTEMV 434
>TC206928 similar to UP|Q9LNC6 (Q9LNC6) F9P14.7 protein (At1g06210/F9P14_4),
partial (58%)
Length = 1459
Score = 102 bits (253), Expect = 7e-22
Identities = 72/270 (26%), Positives = 135/270 (49%), Gaps = 8/270 (2%)
Frame = +1
Query: 5 LVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLET 64
+V+ AT + + P+W +N+ IC ++N D +VVK +K++I +S VQ L+L LLE
Sbjct: 175 MVDEATLETMEEPNWGMNLRICGMINSDQFNGSEVVKAIKRKINHKSPVVQTLSLDLLEA 354
Query: 65 IIKNCGDIVHMHVAERDVLHEMVKIV-KKKPDPRVREKIMILIDTWQEAFGGPRARYPQY 123
NC D V +A VL E+++++ + + R + LI W E+ A P +
Sbjct: 355 CAMNC-DKVFSEIASEKVLDEIIRLIDNPQAHHQTRSRAFQLIRAWGES--EDLAYLPVF 525
Query: 124 YAAYQELLHSGAVFPQRSEQSAPVFTPLQTQPLTSYPQNIRDSDAQPHTAESSA--ESEF 181
Y L G P P ++ P+ A+ H + A S +
Sbjct: 526 RQTYMSL--KGRDEPVDMAGGNSPHVPYASESYVDAPERYPIPQAELHDIDDPAAFSSNY 699
Query: 182 PTLSLSEIQN----ARGIMDVLAEMLSAIEPGNKEGLRQEVIVDLVEQCRTYKQRVVHLV 237
+S+ E + AR +++L+ +L++ + L++++ V L+++C+ + +V
Sbjct: 700 QHISVEERKEHLVVARNSLELLSSILNS--DAEPKTLKEDLTVSLLDKCKQSLSIIKGIV 873
Query: 238 NS-TSDESLLCQGLALNDDLQRVLAKHESI 266
S T+DE+ L + L LND+LQ++++K+E +
Sbjct: 874 ESTTNDEATLFEALYLNDELQQIVSKYEEL 963
>TC217788 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like protein
(AT5g16880/F2K13_30), partial (52%)
Length = 1178
Score = 99.0 bits (245), Expect = 6e-21
Identities = 58/129 (44%), Positives = 82/129 (62%), Gaps = 5/129 (3%)
Frame = +1
Query: 5 LVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLET 64
LVE ATS+ L PDWALN+++CD++N D S ++V+G+KKRI +S +VQ LAL LLET
Sbjct: 337 LVEDATSEALDEPDWALNLDLCDLINADKLSSVELVRGIKKRIVLKSPRVQYLALVLLET 516
Query: 65 IIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRV----REKIMILIDTWQEAFGGPRARY 120
++KNC AER VL EMV+++ DP+ R K +++I+ W E+ G RY
Sbjct: 517 LVKNCEKAFSEVAAER-VLDEMVRLI---DDPQTVVNNRNKALMMIEAWAESTG--ELRY 678
Query: 121 -PQYYAAYQ 128
P Y Y+
Sbjct: 679 LPVYEETYK 705
>TC217789 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like protein
(AT5g16880/F2K13_30), partial (29%)
Length = 495
Score = 87.4 bits (215), Expect = 2e-17
Identities = 50/111 (45%), Positives = 72/111 (64%), Gaps = 4/111 (3%)
Frame = +3
Query: 5 LVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLET 64
LVE ATS+ L P+WALN+++CD++N D ++V+G+KKRI +S +VQ LAL LLET
Sbjct: 174 LVEDATSEALDEPEWALNLDLCDLVNTDKLNCVELVRGIKKRIILKSPRVQYLALVLLET 353
Query: 65 IIKNCGDIVHMHVAERDVLHEMVKIVKKKPDPRV----REKIMILIDTWQE 111
++KNC AER VL EMVK++ DP+ R K +++I+ E
Sbjct: 354 LVKNCEKAFSEVAAER-VLDEMVKLI---DDPQTVVNNRNKALMMIEAXGE 494
>AW759150 similar to GP|21430672|gb| RE65163p {Drosophila melanogaster},
partial (3%)
Length = 250
Score = 79.3 bits (194), Expect = 5e-15
Identities = 46/79 (58%), Positives = 51/79 (64%), Gaps = 8/79 (10%)
Frame = +3
Query: 394 SVNNQP---TNVAVPNSPLSPQFQQQQTFI-SQGVYYPNGSMPSVGSPQYEQSRYAQSTG 449
SVN+QP TNVA SP PQFQQQQT I SQ +YPN + P+ SPQY+QS Y QST
Sbjct: 12 SVNSQPPQPTNVAGQTSPYGPQFQQQQTVITSQSRFYPNKNTPNTCSPQYKQSLYTQSTS 191
Query: 450 PAWNGQAA----QQQQPPS 464
P NGQ Q QQPPS
Sbjct: 192 PT*NGQVTQXQQQHQQPPS 248
>BE661161
Length = 718
Score = 72.0 bits (175), Expect = 8e-13
Identities = 37/109 (33%), Positives = 61/109 (55%), Gaps = 1/109 (0%)
Frame = +2
Query: 5 LVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALTLLET 64
+V+ AT + + P+W +N+ IC ++N D +VVK +K++I +S VQ L+L LLE
Sbjct: 209 MVDEATLETMEEPNWGMNLRICSMINSDQFNGSEVVKAIKRKINHKSPVVQALSLDLLEA 388
Query: 65 IIKNCGDIVHMHVAERDVLHEMVKIV-KKKPDPRVREKIMILIDTWQEA 112
NC D V +A VL EM++++ + + R + LI W E+
Sbjct: 389 CAMNC-DKVFSEIASEKVLDEMIRLIDNPQAQHQTRSRAFQLIRAWGES 532
>BG363401
Length = 405
Score = 71.2 bits (173), Expect = 1e-12
Identities = 33/74 (44%), Positives = 47/74 (62%)
Frame = +3
Query: 1 MVNPLVERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIGSRSSKVQLLALT 60
M LV ATS+ L DW NIEIC+++ D Q++D VK +KKR+GS+ QL A+
Sbjct: 147 MAAELVNGATSEKLAETDWTKNIEICELVAHDKRQARDAVKAIKKRLGSKHPNTQLFAVM 326
Query: 61 LLETIIKNCGDIVH 74
LLE ++ N G+ +H
Sbjct: 327 LLEMLMNNIGEHIH 368
>BU082307
Length = 416
Score = 64.7 bits (156), Expect = 1e-10
Identities = 28/33 (84%), Positives = 31/33 (93%)
Frame = +3
Query: 1 MVNPLVERATSDMLIGPDWALNIEICDILNRDP 33
MVN +VERATSDMLIGPDWA+NIEICD+LN DP
Sbjct: 189 MVNSMVERATSDMLIGPDWAMNIEICDMLNHDP 287
>BE474140
Length = 427
Score = 58.9 bits (141), Expect = 7e-09
Identities = 35/85 (41%), Positives = 44/85 (51%)
Frame = +3
Query: 35 QSKDVVKGLKKRIGSRSSKVQLLALTLLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKP 94
Q+KD +K LKKR+ S++ ++QLLAL P
Sbjct: 270 QAKDAIKILKKRLSSKNPQIQLLALF---------------------------------P 350
Query: 95 DPRVREKIMILIDTWQEAFGGPRAR 119
D RVREKI+ILIDTWQEAFGGP +
Sbjct: 351 DLRVREKILILIDTWQEAFGGPSGK 425
Score = 58.2 bits (139), Expect = 1e-08
Identities = 26/42 (61%), Positives = 32/42 (75%)
Frame = +1
Query: 7 ERATSDMLIGPDWALNIEICDILNRDPAQSKDVVKGLKKRIG 48
ERATSDMLIGPDWA+NIE+CDI+N DP + + G K +G
Sbjct: 28 ERATSDMLIGPDWAINIELCDIINMDPREHHVLGTGSKFIVG 153
>TC219976 similar to UP|OAZ_HUMAN (P54368) Ornithine decarboxylase antizyme
(ODC-Az), partial (6%)
Length = 1046
Score = 53.9 bits (128), Expect = 2e-07
Identities = 24/53 (45%), Positives = 38/53 (71%)
Frame = +3
Query: 217 EVIVDLVEQCRTYKQRVVHLVNSTSDESLLCQGLALNDDLQRVLAKHESIASG 269
E +DLV QC KQRV+HLV ++ DE ++ + + LN+ LQ+VLA+H+ + +G
Sbjct: 3 EFTLDLV*QCSFQKQRVMHLVMASRDERIVSRAIELNEQLQKVLARHDDLLAG 161
>BF425076 similar to GP|28381653|gb| CG1693-PA {Drosophila melanogaster},
partial (1%)
Length = 408
Score = 52.0 bits (123), Expect = 8e-07
Identities = 25/32 (78%), Positives = 26/32 (81%), Gaps = 2/32 (6%)
Frame = +1
Query: 440 EQSRYAQSTGPAWNGQAA--QQQQPPSPVNGA 469
EQS Y Q+TGPAWNGQ A QQQQPPSPV GA
Sbjct: 1 EQSLYTQNTGPAWNGQVAQQQQQQPPSPVYGA 96
Score = 30.4 bits (67), Expect = 2.6
Identities = 13/18 (72%), Positives = 14/18 (77%)
Frame = +3
Query: 466 VNGAQSGGSFPPPPWEAQ 483
+ G QSG S PPPPWEAQ
Sbjct: 354 LTGTQSG-SLPPPPWEAQ 404
>TC210294
Length = 769
Score = 45.1 bits (105), Expect = 1e-04
Identities = 18/40 (45%), Positives = 31/40 (77%)
Frame = +3
Query: 230 KQRVVHLVNSTSDESLLCQGLALNDDLQRVLAKHESIASG 269
KQRV+HLV ++ DE ++ + + LN+ LQ+VLA+H+ + +G
Sbjct: 3 KQRVMHLVMASRDERIISRAIELNEQLQKVLARHDDLLAG 122
>TC204718 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;} , partial (5%)
Length = 1417
Score = 45.1 bits (105), Expect = 1e-04
Identities = 63/263 (23%), Positives = 77/263 (28%), Gaps = 36/263 (13%)
Frame = +2
Query: 408 PLSPQFQQQQTFISQGVY----------YPNGSMPSVGSPQYEQSRYAQSTGPA------ 451
P +P Q ++ G Y P P S Y + + QST P
Sbjct: 20 PAAPMQQSGYGYVQPGAYSGPPQYNVPQQPYAGYPPQSSGGYSAANWDQSTAPQQQSAHA 199
Query: 452 ---WNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQMVVT 508
+ Q QQQ P P +G S PP +Q + + Y P Q
Sbjct: 200 GYDYYSQQQQQQNPAPPSDGTAYNYSQPPSSGYSQPGQGYAQDGYGGYHAPPQ------- 358
Query: 509 HVQGAPLPQGPQAMGYDQAVGMYMQQPNASHMSAMNNQVQSNQF--GMHPQ--YAQGVAA 564
G P P Q GY A H + +Q S Q P Y G
Sbjct: 359 SGYGQP-PSYDQQQGYSSAASYGSNPAPEGHTANYGSQGDSTQAPPAQPPSQGYGTGQQP 535
Query: 565 SPYMGMVPHQMQNGPVAHPMYAQQMYGNQ------YMGGYG-------YGQQPQQGVQYV 611
SP Q Q G P YG+Q Y GYG G P G
Sbjct: 536 SPNAANYAPQAQPGYGVPPTSQPATYGSQPPAQSGYGSGYGPPQTQKPSGTPPVYGQSQS 715
Query: 612 EQQMHGLSMSDDRNSSYVPPKKP 634
G S S Y P + P
Sbjct: 716 PNTAGGYGQSGHLQSGYPPSQPP 784
Score = 33.5 bits (75), Expect = 0.31
Identities = 54/240 (22%), Positives = 71/240 (29%), Gaps = 15/240 (6%)
Frame = +2
Query: 280 NPAPPGALIDVGGPLVDTGDTSKQSDERSPSGAEGGSQTLN----------QLLLPAPPT 329
NPAP G + G GD+++ + PS G Q + Q PPT
Sbjct: 428 NPAPEGHTANYGSQ----GDSTQAPPAQPPSQGYGTGQQPSPNAANYAPQAQPGYGVPPT 595
Query: 330 SN----GSAPPIKIDPKLDLLSGDDYNSPKADNSLALVPV-GQQQPASPMSQQNALVLFD 384
S GS PP + G Y P+ PV GQ Q +
Sbjct: 596 SQPATYGSQPPAQSG------YGSGYGPPQTQKPSGTPPVYGQSQSPNTAG--------- 730
Query: 385 MFSNGSNAPSVNNQPTNVAVPNSPLSPQFQQQQTFISQGVYYPNGSMPSVGSPQYEQSRY 444
G + + P + P+ + Q G Y P G P Y
Sbjct: 731 --GYGQSGHLQSGYPPSQPPPSGGYAQPESGSQKAPPSG--YGGAVQPGYGPPSYG---- 886
Query: 445 AQSTGPAWNGQAAQQQQPPSPVNGAQSGGSFPPPPWEAQAAENGSPVAGNQYPQPLQVTQ 504
G GQ Q PPS N + + PP + G G P Q Q
Sbjct: 887 ----GAPAGGQPGYGQAPPSYSNSSYGAAGYAQPPVYSSDGNVGGTTRGAYDGAPAQPVQ 1054
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.130 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,190,174
Number of Sequences: 63676
Number of extensions: 419348
Number of successful extensions: 2361
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 2151
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2278
length of query: 658
length of database: 12,639,632
effective HSP length: 103
effective length of query: 555
effective length of database: 6,081,004
effective search space: 3374957220
effective search space used: 3374957220
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0487.5


