

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.5
(58 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
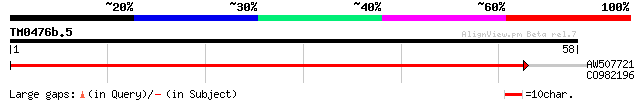
Score E
Sequences producing significant alignments: (bits) Value
AW507721 weakly similar to GP|27764548|gb polyprotein {Glycine m... 40 2e-04
CO982196 25 6.3
>AW507721 weakly similar to GP|27764548|gb polyprotein {Glycine max}, partial
(3%)
Length = 464
Score = 40.4 bits (93), Expect = 2e-04
Identities = 17/53 (32%), Positives = 32/53 (60%)
Frame = +1
Query: 1 KLILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTCLTYQKAKV 53
K+I+ E H S++ H G T+ + F+ P M++ ++++V C+ Q+AKV
Sbjct: 274 KMIMTESHASKVGGHAGTTRTIVRINAQFYWPKMREDIMKFVQECVICQQAKV 432
>CO982196
Length = 812
Score = 25.4 bits (54), Expect = 6.3
Identities = 14/44 (31%), Positives = 21/44 (46%)
Frame = +1
Query: 2 LILDEGHKSRLSIHPGMTKMYQDLKLHFWCPGMKKRVVEYVLTC 45
L+L E S L H G + ++ + + GMKK +YV C
Sbjct: 373 LLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAAC 504
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,726,245
Number of Sequences: 63676
Number of extensions: 26378
Number of successful extensions: 116
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 116
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 116
length of query: 58
length of database: 12,639,632
effective HSP length: 34
effective length of query: 24
effective length of database: 10,474,648
effective search space: 251391552
effective search space used: 251391552
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0476b.5


