

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
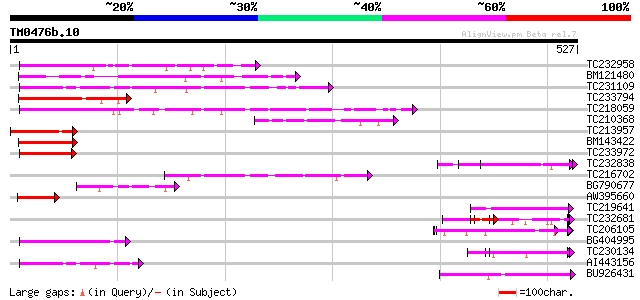
Query= TM0476b.10
(527 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232958 145 4e-35
BM121480 127 1e-29
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 99 5e-21
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 95 9e-20
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 92 4e-19
TC210368 87 2e-17
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 84 1e-16
BM143422 77 2e-14
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 63 3e-10
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 62 5e-10
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 60 2e-09
BG790677 60 2e-09
AW395660 58 9e-09
TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 54 2e-07
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 54 2e-07
TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-in... 53 3e-07
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 53 3e-07
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 52 5e-07
AI443156 52 6e-07
BU926431 51 1e-06
>TC232958
Length = 758
Score = 145 bits (366), Expect = 4e-35
Identities = 102/245 (41%), Positives = 144/245 (58%), Gaps = 21/245 (8%)
Frame = +1
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP +LI EILLRLPV+SL+RFK VC SW FLISDP+FA+SHFDL AA R+L
Sbjct: 61 LPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIASSAP 240
Query: 70 KLESLDL------ESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWN 123
+L S+D +S+S+ VT++LP P K H + +GSCRGF+LL + VWN
Sbjct: 241 ELRSIDFNASLHDDSASVAVTVDLPAP-KPYFH-FVEIIGSCRGFILL--HCLSHLCVWN 408
Query: 124 PSTGFYKQI-LSFSDFMLDSLY-----GFGYDNSTDDYFLVLIGLIWVK---AIIQAFSV 174
P+TG +K + LS F D+++ GFGYD STDD FLV+ K + FS+
Sbjct: 409 PTTGVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDD-FLVVHACYNPKHQANCAEIFSL 585
Query: 175 KTNS------CDFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAY 228
+ N+ F Y + +Y + + + G FLN ++HWL +F + S V++A+
Sbjct: 586 RANAWKGIEGIHFPYTHFRYTN--RYNQFGSFLNGAIHWL------AFRINASINVIVAF 741
Query: 229 DLLEK 233
DL+E+
Sbjct: 742 DLVER 756
>BM121480
Length = 917
Score = 127 bits (319), Expect = 1e-29
Identities = 97/270 (35%), Positives = 130/270 (47%), Gaps = 8/270 (2%)
Frame = +3
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDK 68
TLP ELI EILLRLPV+SLLRFKCV F D+ P +PCL
Sbjct: 63 TLPQELIREILLRLPVKSLLRFKCVL-----------FKTCSRDVVYFPLPLPSIPCL-- 203
Query: 69 NKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNPSTGF 128
+L+ + LGSCRG +LL YD + +I+WNPS G
Sbjct: 204 -RLDDFGIRPK---------------------ILGSCRGLVLLYYDDSANLILWNPSLGR 317
Query: 129 YKQILSFSDFMLDSLYGFGYDNSTDDYFLVLIGL--IWVKAIIQAFSVKTNSCDFKYVNA 186
+K + ++ D + YGFGYD S D+Y L+LIGL + +S KT S +K
Sbjct: 318 HKXLPNYRDDITSFPYGFGYDESKDEYLLILIGLPKFGPETGADIYSFKTES--WKTDTI 491
Query: 187 QYRDLGYHY------RHGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLLEKSLSEIPL 240
Y L + R G LN +LHW V FS V+IA+DL+E++LS IPL
Sbjct: 492 VYDPLXRYXAEDXIARAGSLLNGALHWFV------FSESKXDHVIIAFDLVERTLSXIPL 653
Query: 241 SPELAKPVLTAEGAPKFYHVRVLGGCLSLC 270
LA + A Y ++++G CL +C
Sbjct: 654 --PLADRSTVQKDA--VYGLKIMGXCLXVC 731
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 99.0 bits (245), Expect = 5e-21
Identities = 88/305 (28%), Positives = 148/305 (47%), Gaps = 13/305 (4%)
Frame = +3
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP E++ EIL RLPV+S++R + C W +I F H LN + + +L ++
Sbjct: 93 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFH--LNKSHSSLIL---RHRS 257
Query: 70 KLESLDLESSSLF-VTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNPSTGF 128
L SLDL+S V L+ P C S NS+ LGS G + ++ + + +WNP
Sbjct: 258 HLYSLDLKSPEQNPVELSHPLMCYS---NSIKVLGSSNGLLCIS-NVADDIALWNPFLRK 425
Query: 129 YKQILS------FSDFMLDSLYGFGYDNSTDDYFLVLIGLI------WVKAIIQAFSVKT 176
++ + + S +YGFG+ + ++DY L+ I + +Q +++K+
Sbjct: 426 HRILPADRFHRPQSSLFAARVYGFGHHSPSNDYKLLSITYFVDLQKRTFDSQVQLYTLKS 605
Query: 177 NSCDFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLLEKSLS 236
+S +K + + L GVF++ SLHWLVT D +++++DL ++
Sbjct: 606 DS--WKNLPSMPYALCCARTMGVFVSGSLHWLVTRKLQPHEPD----LIVSFDLTRETFH 767
Query: 237 EIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKGGRRDRAEIWVMKEYKVQSSWTKA 296
E+PL +T G V +LGGC LC R ++WVM+ Y ++SW K
Sbjct: 768 EVPLP-------VTVNGDFDM-QVALLGGC--LCVVEHRGTGFDVWVMRVYGSRNSWEKL 917
Query: 297 FVVTD 301
F + +
Sbjct: 918 FTLLE 932
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 94.7 bits (234), Expect = 9e-20
Identities = 60/117 (51%), Positives = 71/117 (60%), Gaps = 12/117 (10%)
Frame = +1
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDK 68
TLP ELI EILLRLPV+SLLRFKCVC S+ LISDPQF SH+ L A+PTHRL+L D
Sbjct: 106 TLPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAASPTHRLILRSHD- 282
Query: 69 NKLESLDLESSSLFVT---------LNLPP-PCKSRDHNSL--YFLGSCRGFMLLAY 113
+ + + S+F T L LP PC D + LGSCRG +LL Y
Sbjct: 283 --FYAQSIATESVFKTCSRDVVYFPLPLPSIPCLRLDDFGIRPKILGSCRGLVLLYY 447
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 92.4 bits (228), Expect = 4e-19
Identities = 105/407 (25%), Positives = 180/407 (43%), Gaps = 37/407 (9%)
Frame = +3
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDL-NAAPTHRLLLPCLDK 68
LP EL++ +L RLP + LL KCVC SWF LI+DP F +++ + N+ + L + +
Sbjct: 66 LPGELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHLLVIRR 245
Query: 69 NKLESLDLESSSLFVTLNLPPPCKSRD-------HNSLY-----FLGSCRGFMLLAYDYN 116
L S L N P S D +NS + LG C G L + N
Sbjct: 246 PFFSGLKTYISVLSWNTNDPKKHVSSDVLNPPYEYNSDHKYWTEILGPCNGIYFLEGNPN 425
Query: 117 RQVIVWNPSTGFYKQI-----------LSFSDFMLDSLYGFGYDNSTDDYFLVLIGLIWV 165
++ NPS G +K + +F+D+ GFG+D T+DY +V++ +W+
Sbjct: 426 ---VLMNPSLGEFKALPKSHFTSPHGTYTFTDYA-----GFGFDPKTNDYKVVVLKDLWL 581
Query: 166 KAI---------IQAFSVKTNSCDFKYVNAQYRDLGYHY----RHGVFLNNSLHWLVTTT 212
K + +S+ +NS ++ ++ L R + NN HW
Sbjct: 582 KETDEREIGYWSAELYSLNSNS--WRKLDPSLLPLPIEIWGSSRVFTYANNCCHWWGFVE 755
Query: 213 DTSFSNDTSFLVVIAYDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYK 272
++ + D VV+A+D++++S +I + P++ G + G L +
Sbjct: 756 ESDATQD----VVLAFDMVKESFRKIRV-PKIRDSSDEKFGTLVPFEESASIGFLVYPVR 920
Query: 273 GGRRDRAEIWVMKEYKVQSSWTKAFVVTDCDIPCIHFYPIRFIERGGVLGSNGNGRLMTF 332
G + R ++WVMK+Y + SW K + V P++ I + ++G G R
Sbjct: 921 GTEK-RFDVWVMKDYWDEGSWVKQYSVG----------PVQVIYK--LVGFYGTNRFFWK 1061
Query: 333 NAEGKLLEHHKYGQEIKNVRKDLAMYRESEYKNIQIYFEMYRESLLS 379
++ +L+ + + +N R DL +Y K+ I Y ESL+S
Sbjct: 1062DSNERLVLY-----DSENTR-DLQVYG----KHDSIRAAKYTESLVS 1172
>TC210368
Length = 815
Score = 87.0 bits (214), Expect = 2e-17
Identities = 60/138 (43%), Positives = 77/138 (55%), Gaps = 4/138 (2%)
Frame = +1
Query: 228 YDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKGGRRDRAEIWVMKEY 287
+DL+E++LSEIPL LA + A Y +R++GGCLS+C EIWVMKEY
Sbjct: 1 FDLVERTLSEIPLP--LADRSTVQKDA--VYGLRIMGGCLSVCCSSC--GMTEIWVMKEY 162
Query: 288 KVQSSWTKAFVVTDCDIPCIHFYPIRFIERGGVLGSN--GNGRLMTFNAEGKLLEH--HK 343
KV+SSWT F++ + PI I+ G +LGSN G GRL N +G+LLEH
Sbjct: 163 KVRSSWTMTFLIHTSN----RISPICTIKDGEILGSNCAGTGRLEKRNDKGELLEHFMDT 330
Query: 344 YGQEIKNVRKDLAMYRES 361
GQ AMY ES
Sbjct: 331 KGQRFSCANLQAAMYTES 384
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 84.3 bits (207), Expect = 1e-16
Identities = 43/63 (68%), Positives = 48/63 (75%)
Frame = +3
Query: 1 MANEKTNPTLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHR 60
M + TLP ELI EILLR PVRS+LRFKCVC SW LISDPQF +HFDL A+PTHR
Sbjct: 6 MEKHTLSVTLPLELIREILLRSPVRSVLRFKCVCKSWLSLISDPQF--THFDLAASPTHR 179
Query: 61 LLL 63
L+L
Sbjct: 180LIL 188
Score = 38.9 bits (89), Expect = 0.006
Identities = 30/90 (33%), Positives = 37/90 (40%), Gaps = 8/90 (8%)
Frame = +1
Query: 121 VWNPSTGFYKQILSFSDFMLDSL-YGFGYDNSTDDYFLVLI-------GLIWVKAIIQAF 172
+ NP YK + F + YG YD TDD L+ L K IQ F
Sbjct: 217 ILNPIIRVYK*LSDFKVMLTKKFQYGLVYDELTDD*STCLLLFQLFSDSLDECKPEIQVF 396
Query: 173 SVKTNSCDFKYVNAQYRDLGYHYRHGVFLN 202
S KTN +N Y+ LG + G FLN
Sbjct: 397 SFKTNLRSRYNINVPYKHLGLEFGAGSFLN 486
>BM143422
Length = 424
Score = 77.0 bits (188), Expect = 2e-14
Identities = 39/56 (69%), Positives = 44/56 (77%), Gaps = 1/56 (1%)
Frame = +1
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLN-AAPTHRLLL 63
TLP +LI ILL+LPV+S+ RFKCVC SW LISDPQF SHFDL A P+HRLLL
Sbjct: 226 TLPLDLIELILLKLPVKSVTRFKCVCKSWLSLISDPQFGFSHFDLALAVPSHRLLL 393
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 63.2 bits (152), Expect = 3e-10
Identities = 33/53 (62%), Positives = 39/53 (73%)
Frame = -2
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLL 62
LP E+I EILLRLPV+SL FK VC S LIS+P FA+ HF+ NAA T +LL
Sbjct: 540 LPQEIIIEILLRLPVKSLPSFKFVCKS*LSLISNPHFAKWHFERNAAHTEKLL 382
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 62.4 bits (150), Expect = 5e-10
Identities = 35/128 (27%), Positives = 66/128 (51%)
Frame = +2
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKED 457
E NEV+ ++++EE ++ E V+ AED+E+E +D+D D D
Sbjct: 254 EENEVKVLVEEDDEE---------------YESVDEVVDDAEDDEDEEEDDDDDEGDDND 388
Query: 458 ESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDE 517
+ E+ + + G+ +DE+ DEEE + D ++ +S+ +S+ DE+ E++
Sbjct: 389 DEEDDAPDGGDDDDDED---------DEEEGDVQRGGEPDDDDNDSDDDSDDDEDEDEED 541
Query: 518 EEESSEDD 525
EEE E++
Sbjct: 542 EEEQGEEE 565
Score = 59.7 bits (143), Expect = 3e-09
Identities = 28/108 (25%), Positives = 58/108 (52%)
Frame = +2
Query: 418 LHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATE 477
+ + K L+ + +E +V ED+EE + DE D ++DE EE +D E ++++ +
Sbjct: 221 MKATKELSEQVEENEVKVLVEEDDEEYESVDEVVDDAEDDEDEEEDDDDDEGDDNDDEED 400
Query: 478 DVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
D D++++ + EE E + + N ++ ++D+E+E ED+
Sbjct: 401 DAPDGGDDDDDEDDEEEGDVQRGGEPDDDDNDSDDDSDDDEDEDEEDE 544
Score = 52.0 bits (123), Expect = 6e-07
Identities = 30/97 (30%), Positives = 52/97 (52%), Gaps = 7/97 (7%)
Frame = +2
Query: 438 AEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATD 497
++D+E+E EDE+ ++ED E E E+EEA+ D E + EEE +V++ D
Sbjct: 509 SDDDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPEENGEEEEEDVDD-ED 685
Query: 498 HEEVE-------SEKESNKDEEATEDEEEESSEDDYI 527
E+ E S+K+ + D++ ED+E S ++
Sbjct: 686 CEKAEAPPKRKRSDKDDSDDDDGGEDDERPSKR*SWV 796
Score = 39.3 bits (90), Expect = 0.004
Identities = 24/87 (27%), Positives = 45/87 (51%)
Frame = +2
Query: 434 EVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVE 493
+++A ++ E+ E+E +ED+ E S + E +D E DEEE+ + +
Sbjct: 218 KMKATKELSEQVEENEVKVLVEEDDEEYESVD--------EVVDDAEDDEDEEEDDDD-D 370
Query: 494 EATDHEEVESEKESNKDEEATEDEEEE 520
E D+++ E + D++ ED+EEE
Sbjct: 371 EGDDNDDEEDDAPDGGDDDDDEDDEEE 451
Score = 36.6 bits (83), Expect = 0.028
Identities = 26/85 (30%), Positives = 46/85 (53%), Gaps = 10/85 (11%)
Frame = +2
Query: 400 NEVEEATKDEEEEASGLSL---HSVKSL-TCEFKEATEEVEAAE--DEEEETTEDEDA-- 451
++ +E +DEEE+ L + ++ L T E +EA+ + E E +EEEE +DED
Sbjct: 515 DDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPEENGEEEEEDVDDEDCEK 694
Query: 452 --SDDKEDESEEASTEDGEATEDEE 474
+ K S++ ++D + ED+E
Sbjct: 695 AEAPPKRKRSDKDDSDDDDGGEDDE 769
Score = 34.3 bits (77), Expect = 0.14
Identities = 20/72 (27%), Positives = 35/72 (47%), Gaps = 3/72 (4%)
Frame = +2
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATED 516
D +A+ E E E+ E VE +E E EV + + +E E E + + + + +D
Sbjct: 212 DSKMKATKELSEQVEENEVKVLVEEDDEEYESVDEVVDDAEDDEDEEEDDDDDEGDDNDD 391
Query: 517 EEE---ESSEDD 525
EE+ + +DD
Sbjct: 392 EEDDAPDGGDDD 427
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 60.1 bits (144), Expect = 2e-09
Identities = 56/204 (27%), Positives = 91/204 (44%), Gaps = 11/204 (5%)
Frame = +3
Query: 145 GFGYDNSTDDYFLVLIGLIW------VKAIIQAFSVKTNSCDFKYVNAQYRDLGYHYRHG 198
GFG+D+ T DY LV I A ++ ++++ N+ +K + + L G
Sbjct: 135 GFGFDHKTRDYKLVRISYFVDLHDRSFDAQVKLYTLRANA--WKTLPSLPYALCCARTMG 308
Query: 199 VFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLLEKSLSEIPLSPELAKPVLTAEGAPKFY 258
VF+ NSLHW+VT D ++IA+DL E+PL P
Sbjct: 309 VFVGNSLHWVVTRKLEPDQPD----LIIAFDLTHDIFRELPL------PDTGGVDGGFEI 458
Query: 259 HVRVLGGCLSLCYKGGRRDRAEIWVMKEYKVQSSWTKAFVVTDC----DIPCIHFYPIRF 314
+ +LGG L + + R ++WVM+EY + SW K F + + + C+ P+ +
Sbjct: 459 DLALLGGSLCMTVNF-HKTRIDVWVMREYNRRDSWCKVFTLEESREMRSLKCVR--PLGY 629
Query: 315 IERGG-VLGSNGNGRLMTFNAEGK 337
G VL + RL ++ E K
Sbjct: 630 SSDGNKVLLEHDRKRLFWYDLEKK 701
>BG790677
Length = 419
Score = 60.1 bits (144), Expect = 2e-09
Identities = 44/104 (42%), Positives = 55/104 (52%), Gaps = 8/104 (7%)
Frame = +1
Query: 63 LPCLDKNKLESLDLESSSLF----VTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQ 118
L C ++ S+D+ +S F VT L P D F+ SCR F+LL DYNR
Sbjct: 73 LACSSVPEILSIDINASFYFFSDYVT*KLSAPIHFGDR----FVCSCRDFLLL--DYNRS 234
Query: 119 VIVWNPSTGFYKQILSFSDFMLDSL----YGFGYDNSTDDYFLV 158
++NPSTG +K F D+ S YGFGYD STDDY LV
Sbjct: 235 SFIFNPSTGLHK----FVDWSPISNNMGGYGFGYDPSTDDYLLV 354
>AW395660
Length = 381
Score = 58.2 bits (139), Expect = 9e-09
Identities = 26/39 (66%), Positives = 30/39 (76%)
Frame = +3
Query: 8 PTLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQF 46
P LP EL+ EIL RLPV+SLL+F+CVC SW LI DP F
Sbjct: 258 PFLPDELVVEILSRLPVKSLLQFRCVCKSWMSLIYDPYF 374
>TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 942
Score = 53.9 bits (128), Expect = 2e-07
Identities = 26/96 (27%), Positives = 51/96 (53%)
Frame = +1
Query: 429 KEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEE 488
K+A++ E ++EE+ D+DA D ++E +E + +GE D E + + +
Sbjct: 370 KDASDSDEDGDEEED----DDDADDQDDEEGDEDFSGEGEEEADPEDDPEANGAGGSDSD 537
Query: 489 ATEVEEATDHEEVESEKESNKDEEATEDEEEESSED 524
+ ++ D ++ ++ + +DEE EDE+EE ED
Sbjct: 538 GDDDDDGGDDDDDDNGDDDGEDEEEGEDEDEEDEED 645
Score = 38.9 bits (89), Expect = 0.006
Identities = 22/85 (25%), Positives = 45/85 (52%), Gaps = 7/85 (8%)
Frame = +1
Query: 448 DEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVE----- 502
++DASD S EDG+ ED++ +D + +E+ + E EE D E+
Sbjct: 367 NKDASD---------SDEDGDEEEDDDDADDQDDEEGDEDFSGEGEEEADPEDDPEANGA 519
Query: 503 --SEKESNKDEEATEDEEEESSEDD 525
S+ + + D++ +D+++++ +DD
Sbjct: 520 GGSDSDGDDDDDGGDDDDDDNGDDD 594
Score = 32.7 bits (73), Expect = 0.41
Identities = 22/104 (21%), Positives = 47/104 (45%), Gaps = 6/104 (5%)
Frame = +1
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDE------D 450
K+A++ +E +EE++ + E +E + E EEE ED+
Sbjct: 370 KDASDSDEDGDEEEDDDDADD---------QDDEEGDEDFSGEGEEEADPEDDPEANGAG 522
Query: 451 ASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEE 494
SD D+ ++ +D + D++ ++ E ++EE+ +V +
Sbjct: 523 GSDSDGDDDDDGGDDDDDDNGDDDGEDEEEGEDEDEEDEEDVPQ 654
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 53.9 bits (128), Expect = 2e-07
Identities = 31/102 (30%), Positives = 51/102 (49%), Gaps = 10/102 (9%)
Frame = +1
Query: 433 EEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATED--VEATVDEEEEAT 490
++ E ED++E+ D DD EDE EE+ + G +D++ +D E DEEE+
Sbjct: 220 DDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQGE 399
Query: 491 EVEEATDH--------EEVESEKESNKDEEATEDEEEESSED 524
E + T++ EE E+ + +E E+EE+ ED
Sbjct: 400 EADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVDDED 525
Score = 50.8 bits (120), Expect = 1e-06
Identities = 34/137 (24%), Positives = 67/137 (48%), Gaps = 16/137 (11%)
Frame = +1
Query: 403 EEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEA 462
++ +DEE++ + E +E +V+ + +++ +D+D +D+EDE E+
Sbjct: 217 DDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQG 396
Query: 463 STED---------GEATEDEEATEDVEATVDEEEEATEVEEATDHEEVE-------SEKE 506
D E+EEA+ D E + EEE +V++ D E+ E S+K+
Sbjct: 397 EEADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVDD-EDGEKSEAPPKRKRSDKD 573
Query: 507 SNKDEEATEDEEEESSE 523
+ D++ ED++E S+
Sbjct: 574 DSDDDDGGEDDDERPSK 624
Score = 43.5 bits (101), Expect(2) = 6e-05
Identities = 21/79 (26%), Positives = 46/79 (57%)
Frame = +1
Query: 447 EDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKE 506
+D+D ++D+ED+ E+ + + G+ +DE+ EEE ++V+ + ++ +++ +
Sbjct: 211 DDDDDNEDEEDDDEDDAPDGGDDDDDED-----------EEEESDVQRGGEPDDDDNDDD 357
Query: 507 SNKDEEATEDEEEESSEDD 525
++E EDEEE+ E D
Sbjct: 358 DEDEDE--EDEEEQGEEAD 408
Score = 37.7 bits (86), Expect = 0.013
Identities = 25/84 (29%), Positives = 47/84 (55%), Gaps = 3/84 (3%)
Frame = +1
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSL-TCEFKEATEEVEAAE--DEEEETTEDEDASDD 454
+ +E EE +++ EEA + + ++ L T E +EA+ + E E +EEEE +DED +
Sbjct: 358 DEDEDEEDEEEQGEEADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVDDEDG--E 531
Query: 455 KEDESEEASTEDGEATEDEEATED 478
K + + D + ++D++ ED
Sbjct: 532 KSEAPPKRKRSDKDDSDDDDGGED 603
Score = 33.9 bits (76), Expect = 0.18
Identities = 19/54 (35%), Positives = 31/54 (57%)
Frame = +2
Query: 474 EATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDDYI 527
+AT+++ V+E E V+E D EE ESE +E +D E++ +DDY+
Sbjct: 68 KATKELSQHVEENEVTVLVDEEED-EEYESE------DEVVDDGEDDEDDDDYL 208
Score = 33.1 bits (74), Expect = 0.31
Identities = 18/45 (40%), Positives = 28/45 (62%)
Frame = +2
Query: 430 EATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEE 474
+AT+E+ + EE E T D +D+E ESE+ +DGE ED++
Sbjct: 68 KATKEL-SQHVEENEVTVLVDEEEDEEYESEDEVVDDGEDDEDDD 199
Score = 21.2 bits (43), Expect(2) = 6e-05
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +2
Query: 429 KEATEEVEAAEDEEEETTEDEDASDD 454
+E EE E+ EDE + ED++ DD
Sbjct: 128 EEEDEEYES-EDEVVDDGEDDEDDDD 202
>TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-infected
erythrocyte surface antigen, partial (5%)
Length = 858
Score = 53.1 bits (126), Expect = 3e-07
Identities = 41/138 (29%), Positives = 69/138 (49%), Gaps = 9/138 (6%)
Frame = +1
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEE-ETTEDEDASD 453
G KEA +EE + E E + + E E +EVE E+++E E TE++ ++
Sbjct: 424 GKKEAEVIEEKKEVEVTEEKKEIEVTEEKKEAEVIEEKKEVEVTEEKKEIEVTEEKKEAE 603
Query: 454 DKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEK-------- 505
KE++ E TE+ + E E ++ E V+E++E VE + +EVE +
Sbjct: 604 VKEEKKEGEVTEEKKEVEVTEEKKEAEVIVEEKKE---VEVTEEKKEVEVTEGKKEVEVI 774
Query: 506 ESNKDEEATEDEEEESSE 523
E K+ E TE+++E E
Sbjct: 775 EEKKETEVTEEKKEVEVE 828
Score = 46.6 bits (109), Expect = 3e-05
Identities = 38/117 (32%), Positives = 56/117 (47%), Gaps = 2/117 (1%)
Frame = +1
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAED--EEEETTEDEDASDD 454
KEA +EE + E E + + E KE +E E E+ E E T E ++A
Sbjct: 511 KEAEVIEEKKEVEVTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKKEAEVI 690
Query: 455 KEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDE 511
E++ E TE+ + E E ++VE V EE++ TEV E EVE +E + E
Sbjct: 691 VEEKKEVEVTEEKKEVEVTEGKKEVE--VIEEKKETEVTEEKKEVEVEVREEKKESE 855
Score = 43.9 bits (102), Expect = 2e-04
Identities = 44/136 (32%), Positives = 66/136 (48%), Gaps = 7/136 (5%)
Frame = +1
Query: 396 FKEANEV----EEATKDEEEEASGLSLHSVKS--LTCEFKEATEEVEAAEDEEE-ETTED 448
FKE V EA K +E L ++ + LT E ++ AAE +EE E TE
Sbjct: 247 FKEETNVVGDLPEAQKKALDELKKLVQEALNNHELTAPKPEPEKKKPAAEKKEEVEVTEG 426
Query: 449 EDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESN 508
+ ++ E++ E TE E E E E EA V EE++ EVE + +E+E +E
Sbjct: 427 KKEAEVIEEKKEVEVTE--EKKEIEVTEEKKEAEVIEEKK--EVEVTEEKKEIEVTEEKK 594
Query: 509 KDEEATEDEEEESSED 524
+ E E +E E +E+
Sbjct: 595 EAEVKEEKKEGEVTEE 642
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 53.1 bits (126), Expect = 3e-07
Identities = 36/103 (34%), Positives = 48/103 (45%)
Frame = +1
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP E++ EIL RLPVRSLLRF+ SW LI H + L +
Sbjct: 40 LPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLASNTSLILRVDS 219
Query: 70 KLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLA 112
L + + V+LN P C S NS+ LGSC G + ++
Sbjct: 220 DLYQTNFPTLDPPVSLNHPLMCYS---NSITLLGSCNGLLCIS 339
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 52.4 bits (124), Expect = 5e-07
Identities = 33/104 (31%), Positives = 58/104 (55%), Gaps = 6/104 (5%)
Frame = +2
Query: 426 CEFKEATEEVEAAEDEEEETTED-----EDASDDKEDESEEASTEDGEATEDEEATEDVE 480
C+ K +EE + A D E++ +D ED DD ED+ E+ S +DG E+ ++ +D E
Sbjct: 233 CKGKHTSEENKDASDTEDDDDDDDVNDGEDGDDDDEDD-EDFSGDDGG--EEADSDDDPE 403
Query: 481 ATVD-EEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSE 523
A E ++ E ++ D + E + + + D+E EDE+EE+ +
Sbjct: 404 ANGGGESDDDDEDDDDDDDDNDEDDGDEDDDDEEDEDEDEETPQ 535
Score = 48.1 bits (113), Expect = 9e-06
Identities = 25/87 (28%), Positives = 49/87 (55%), Gaps = 8/87 (9%)
Frame = +2
Query: 447 EDEDASDDKEDESEEASTEDGEATEDEEATEDV-------EATVDEEEEATEVEEATDHE 499
E++DASD ++D+ ++ + + +D+E ED EA D++ EA E+ D +
Sbjct: 257 ENKDASDTEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEADSDDDPEANGGGESDDDD 436
Query: 500 EVESEKESNKDE-EATEDEEEESSEDD 525
E + + + + DE + ED+++E ED+
Sbjct: 437 EDDDDDDDDNDEDDGDEDDDDEEDEDE 517
Score = 43.1 bits (100), Expect = 3e-04
Identities = 19/83 (22%), Positives = 44/83 (52%)
Frame = +2
Query: 443 EETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVE 502
+ T+E+ + D ED+ ++ DGE +D++ ++ + D EEA ++ + E
Sbjct: 242 KHTSEENKDASDTEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEADSDDDPEANGGGE 421
Query: 503 SEKESNKDEEATEDEEEESSEDD 525
S+ + D++ +D +E+ ++D
Sbjct: 422 SDDDDEDDDDDDDDNDEDDGDED 490
>AI443156
Length = 414
Score = 52.0 bits (123), Expect = 6e-07
Identities = 40/117 (34%), Positives = 61/117 (51%), Gaps = 2/117 (1%)
Frame = +3
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP E++ EIL RLPV+S++R + C W +I F H LN + T +L ++
Sbjct: 90 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFH--LNKSHTSLIL---RHRS 254
Query: 70 KLESLDLES--SSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNP 124
+L SLDL+S L+ P C S NS+ LGS G + ++ + + +WNP
Sbjct: 255 QLYSLDLKSLLDPNPFELSHPLMCYS---NSIKVLGSSNGLLCIS-NVXDDIALWNP 413
>BU926431
Length = 457
Score = 50.8 bits (120), Expect = 1e-06
Identities = 38/135 (28%), Positives = 61/135 (45%), Gaps = 8/135 (5%)
Frame = +3
Query: 400 NEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEE--------ETTEDEDA 451
+EV + K EEEE + K E +E E E+ E ++ E EDE+
Sbjct: 42 DEVNQNEKHEEEEEDDNVEDAYKHDHNEHEEEGNEHESEEKGDKRGVRGGGSEQEEDENK 221
Query: 452 SDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDE 511
S++ ED + D E E+++ V+ DEE E EE +D +E E+ ++ KD
Sbjct: 222 SNEVED---QRGGGDDEIDENDQEQSAVDTDHDEEFTDEEKEEESDEKEKENSRDEEKDG 392
Query: 512 EATEDEEEESSEDDY 526
E+ E++Y
Sbjct: 393 SVENHNSHEAREENY 437
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,325,799
Number of Sequences: 63676
Number of extensions: 388601
Number of successful extensions: 4371
Number of sequences better than 10.0: 371
Number of HSP's better than 10.0 without gapping: 2482
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3493
length of query: 527
length of database: 12,639,632
effective HSP length: 102
effective length of query: 425
effective length of database: 6,144,680
effective search space: 2611489000
effective search space used: 2611489000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0476b.10


