

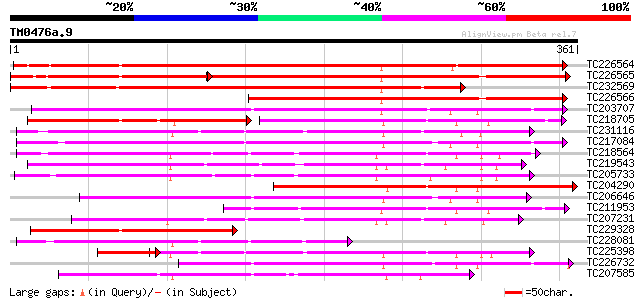
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.9
(361 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226564 387 e-108
TC226565 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif lipase/... 232 e-105
TC232569 302 1e-82
TC226566 203 9e-53
TC203707 190 1e-48
TC218705 weakly similar to GB|AAO63389.1|28950931|BT005325 At1g7... 111 1e-48
TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 182 2e-46
TC217084 179 2e-45
TC218564 176 2e-44
TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase E... 171 4e-43
TC205733 169 2e-42
TC204290 167 9e-42
TC206646 155 2e-38
TC211953 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/... 149 3e-36
TC207231 similar to UP|Q94CH6 (Q94CH6) Family II lipase EXL3, pa... 147 6e-36
TC229328 144 6e-35
TC228081 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 132 3e-31
TC225398 similar to UP|O04320 (O04320) Proline-rich protein APG ... 113 8e-31
TC226732 129 2e-30
TC207585 similar to UP|Q9AYM7 (Q9AYM7) CPRD47 protein, partial (... 129 2e-30
>TC226564
Length = 1337
Score = 387 bits (995), Expect = e-108
Identities = 203/362 (56%), Positives = 257/362 (70%), Gaps = 9/362 (2%)
Frame = +3
Query: 3 CESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLP 62
CE+K+WLV+ LV +VA+ MQH V G SQVPC+F+FGDS+SDSGNNN LPT++K+NF P
Sbjct: 51 CETKSWLVM-FLVFLVANCMQHC-VHGVSQVPCLFIFGDSMSDSGNNNELPTTSKSNFRP 224
Query: 63 YGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRK 122
YGIDFP PTGRY+NGR ID I Q LGF++FIPPFAN +GSDILKGVNYASG +GIR
Sbjct: 225 YGIDFPLG-PTGRYTNGRTEIDIITQFLGFEKFIPPFANTSGSDILKGVNYASGGSGIRN 401
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFL 182
E+G G + LGLQL +HR IVS IA +LG D A QYL +CLYYVNIG+ND+ NYFL
Sbjct: 402 ETGWHYGAAIGLGLQLANHRVIVSEIATKLGSPDLARQYLEKCLYYVNIGSNDYMGNYFL 581
Query: 183 PNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL----VNG 238
P + TS +YT +++ + LI++LS LQ LH GARK L GL +GC P ++ NG
Sbjct: 582 PPFYPTSTIYTIEEFTQVLIEELSLNLQALHDIGARKYALAGLGLIGCTPGMVSAHGTNG 761
Query: 239 SCVEEKNVATFLFNDQLKSLVDRLNKKI-LMDSKFIFINSTAI---IHDKSVGFTVTHHG 294
SC EE+N+A F FN++LK+ VD+ N +SKFIFIN+ A+ + DK GF V
Sbjct: 762 SCAEEQNLAAFNFNNKLKARVDQFNNDFYYANSKFIFINTQALAIELRDK-YGFPVPETP 938
Query: 295 CCPTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNS-SNPAIAYPTNIKH 353
CC G+C+ D PC NR++YVF+D H TE NL+ A+TSYNS +N A YP +IKH
Sbjct: 939 CCLPGLTGECVPDQEPCYNRNDYVFFDAFHPTEQWNLLNALTSYNSTTNSAFTYPMDIKH 1118
Query: 354 LI 355
L+
Sbjct: 1119LV 1124
>TC226565 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif
lipase/hydrolase-like protein, partial (25%)
Length = 1448
Score = 232 bits (591), Expect(2) = e-105
Identities = 121/237 (51%), Positives = 160/237 (67%), Gaps = 5/237 (2%)
Frame = +3
Query: 126 SQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNV 185
S LG ++ GLQL +HR IVS+IA RLG D A QYL +CLYYVNIG+ND+ NYFLP +
Sbjct: 504 SHLGATISFGLQLANHRVIVSQIASRLGSSDLALQYLEKCLYYVNIGSNDYMNNYFLPQL 683
Query: 186 FNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL----VNGSCV 241
+ SR+Y+ +QYA+ALI++LS L LH GARK VL L R+GC P ++ NGSCV
Sbjct: 684 YPASRIYSLEQYAQALIEELSLNLLALHDLGARKYVLARLGRIGCTPSVMHSHGTNGSCV 863
Query: 242 EEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSVGFTVTHHGCCPTNEK 301
EE+N AT +N++LK+LVD+ N + +SKFI I + + D + GF V+ CCP+
Sbjct: 864 EEQNAATSDYNNKLKALVDQFNDRFSANSKFILIPNESNAIDIAHGFLVSDAACCPSG-- 1037
Query: 302 GKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSS-NPAIAYPTNIKHLIQS 357
C D PC NR +Y+FWD +H TEA NLV AI+ YNS+ PA YP +IK L++S
Sbjct: 1038--CNPDQKPCNNRSDYLFWDEVHPTEAWNLVNAISVYNSTIGPAFNYPMDIKQLVES 1202
Score = 167 bits (423), Expect(2) = e-105
Identities = 85/129 (65%), Positives = 102/129 (78%)
Frame = +2
Query: 1 MGCESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANF 60
M CE+KTWLV+ LL + A+ +Q V G SQVPC+F+FGDSLSDSGNNN LPTSAK+N+
Sbjct: 137 MACETKTWLVMVLLF-LAANYLQ-DCVHGVSQVPCLFIFGDSLSDSGNNNELPTSAKSNY 310
Query: 61 LPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGI 120
PYGIDFP PTGR++NGR ID I Q+LGF++FIPPFAN +GS+ILKGVNYASG AGI
Sbjct: 311 RPYGIDFPLG-PTGRFTNGRTEIDIITQLLGFEKFIPPFANTSGSNILKGVNYASGGAGI 487
Query: 121 RKESGSQLG 129
R E+ G
Sbjct: 488 RVETQLAFG 514
>TC232569
Length = 974
Score = 302 bits (774), Expect = 1e-82
Identities = 161/295 (54%), Positives = 207/295 (69%), Gaps = 5/295 (1%)
Frame = +2
Query: 1 MGCESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANF 60
M ++K WL LSL ++ + Q V GESQVPC FV GDSLSD+GNNNNL T+A +N+
Sbjct: 98 MEAKTKPWLALSLFLLATNCMQQR--VHGESQVPCTFVLGDSLSDNGNNNNLQTNASSNY 271
Query: 61 LPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGI 120
PYGID+P T PTGR++NG+N ID I++ LGF E IPP AN +GSDILKG NYASG+AGI
Sbjct: 272 RPYGIDYP-TGPTGRFTNGKNIIDFISEYLGFTEPIPPNANTSGSDILKGANYASGAAGI 448
Query: 121 RKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNY 180
+SG LG N++LG Q+ +HRA +++I RLGG +A +YL +CLYYVNIG+ND+ NY
Sbjct: 449 PFKSGKHLGDNIHLGEQIRNHRATITKIVRRLGGSGRAREYLKKCLYYVNIGSNDYINNY 628
Query: 181 FLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL----V 236
FLP + TSR YT ++Y LI+Q S ++ LH GARK +VGL +GC P +
Sbjct: 629 FLPQFYPTSRTYTLERYTDILIKQYSDDIKKLHRSGARKFAIVGLGLIGCTPNAISRRGT 808
Query: 237 NGS-CVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSVGFTV 290
NG CV E N A FLF+++LKS VD+ K DSKF F+NSTA D+S+GFTV
Sbjct: 809 NGEVCVAELNNAAFLFSNKLKSQVDQF-KNTFPDSKFSFVNSTAGALDESLGFTV 970
>TC226566
Length = 973
Score = 203 bits (517), Expect = 9e-53
Identities = 104/208 (50%), Positives = 140/208 (67%), Gaps = 5/208 (2%)
Frame = +2
Query: 153 GGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTL 212
G D A QYL +CLYY+NIG ND+ NYF P ++ SR+Y+ +QYA+ALI++LS LQ L
Sbjct: 17 GSPDLARQYLEKCLYYLNIGNNDYMGNYFRPQLYPASRIYSLEQYAQALIEELSLNLQAL 196
Query: 213 HHFGARKSVLVGLDRLGCIPKLL----VNGSCVEEKNVATFLFNDQLKSLVDRLNKKILM 268
H GARK VL GL +GC P ++ NGSCVEE N AT+ +N++LK+LVD+ N +
Sbjct: 197 HDLGARKYVLAGLGLIGCTPAVMHSHGTNGSCVEEHNAATYDYNNKLKALVDQFNNRFSA 376
Query: 269 DSKFIFINSTAIIHDKSVGFTVTHHGCCPTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEA 328
+SKFI I++ + D + GF V+ CCP+ C + PC NR +YVFWD +H TEA
Sbjct: 377 NSKFILIHNGSNALDIAHGFLVSDAACCPSG----CNPNQKPCNNRSDYVFWDEVHPTEA 544
Query: 329 ANLVTAITSYNSS-NPAIAYPTNIKHLI 355
NLV AI++YNS+ +PA YP NIK L+
Sbjct: 545 WNLVNAISAYNSTIDPAFTYPMNIKQLV 628
>TC203707
Length = 1394
Score = 190 bits (482), Expect = 1e-48
Identities = 119/354 (33%), Positives = 177/354 (49%), Gaps = 13/354 (3%)
Frame = +1
Query: 15 VVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTG 74
+V++ ++ + G FVFGDSL D+GNNN L T+A+A+ PYGID+P PTG
Sbjct: 49 IVLLGIVLVVVRLKGAEAQRAFFVFGDSLVDNGNNNFLATTARADAPPYGIDYPTGRPTG 228
Query: 75 RYSNGRNPIDKIAQMLGFQEFIPPF-ANLNGSDILKGVNYASGSAGIRKESGSQLGHNVN 133
R+SNG N D I+Q LG + +P L+G +L G N+AS GI ++G Q + +
Sbjct: 229 RFSNGYNIPDFISQSLGAESTLPYLDPELDGERLLVGANFASAGIGILNDTGIQFVNIIR 408
Query: 134 LGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYT 193
+ QL + R++ +G ++ + +N L + +G NDF NY+L SR Y
Sbjct: 409 IYRQLEYWEEYQQRVSGLIGP-EQTERLINGALVLITLGGNDFVNNYYLVPYSARSRQYN 585
Query: 194 PKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL----VNGSCVEEKNVATF 249
Y K +I + L+ L+ GAR+ ++ G LGC+P L NG C E A
Sbjct: 586 LPDYVKYIISEYKKVLRRLYEIGARRVLVTGTGPLGCVPAELAQRSTNGDCSAELQQAAA 765
Query: 250 LFNDQLKSLVDRLNKKILMDSKFIFINSTA-----IIHDKSVGFTVTHHGCC---PTNEK 301
LFN QL ++ +LN +I + F+ +N+ I + + GF + CC P N
Sbjct: 766 LFNPQLVQIIRQLNSEI-GSNVFVGVNTQQMHIDFISNPQRYGFVTSKVACCGQGPYNGL 942
Query: 302 GKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLI 355
G C C NR Y FWD H TE AN + + S YP N+ ++
Sbjct: 943 GLCTPASNLCPNRDSYAFWDPFHPTERANRI-IVQQILSGTSEYMYPMNLSTIM 1101
>TC218705 weakly similar to GB|AAO63389.1|28950931|BT005325 At1g71250
{Arabidopsis thaliana;} , partial (78%)
Length = 1324
Score = 111 bits (278), Expect(2) = 1e-48
Identities = 67/208 (32%), Positives = 106/208 (50%), Gaps = 13/208 (6%)
Frame = +1
Query: 160 QYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARK 219
++L + + V G+ND+ NY LP ++ +SR YT + + L+ + LH G RK
Sbjct: 517 KFLAKSIAVVVTGSNDYINNYLLPGLYGSSRNYTAQDFGNLLVNSYVRQILALHSVGLRK 696
Query: 220 SVLVGLDRLGCIPKLLV-----NGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIF 274
L G+ LGCIP L G CV+ N FN+ L+S+VD+LN+ ++ F++
Sbjct: 697 FFLAGIGPLGCIPTLRAAALAPTGRCVDLVNQMVGTFNEGLRSMVDQLNRN-HPNAIFVY 873
Query: 275 INSTAIIHD-----KSVGFTVTHHGCCPT-NEKGK--CIRDGIPCQNRHEYVFWDGIHTT 326
N+ + D + F V CC +G+ C+ PC +R++YVFWD H T
Sbjct: 874 GNTYRVFGDILNNPAAFAFNVVDRACCGIGRNRGQLTCLPLQFPCTSRNQYVFWDAFHPT 1053
Query: 327 EAANLVTAITSYNSSNPAIAYPTNIKHL 354
E+A V A N + P +YP N++ +
Sbjct: 1054ESATYVFAWRVVNGA-PDDSYPINMQQM 1134
Score = 99.8 bits (247), Expect(2) = 1e-48
Identities = 64/146 (43%), Positives = 90/146 (60%), Gaps = 3/146 (2%)
Frame = +2
Query: 12 SLLVVMVASIMQHSTVLGESQ-VPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPAT 70
++LV+++ S + V +SQ V +FVFGDSL + GNNN L T A+AN+ PYGIDF +
Sbjct: 71 TVLVLVLCSSYGIAEVKSQSQKVSGLFVFGDSLVEVGNNNFLNTIARANYFPYGIDFSKS 250
Query: 71 FPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLN--GSDILKGVNYASGSAGIRKESGSQL 128
TGR+SNG++ ID I +LG PFA+ + G+ IL GVNYAS SAGI ESG
Sbjct: 251 -STGRFSNGKSLIDFIGDLLGIPS-PTPFADPSTVGTRILYGVNYASASAGILDESGRHY 424
Query: 129 GHNVNLGLQLLHHRAIVSRIAHRLGG 154
G +L Q+L+ +++ + G
Sbjct: 425 GDRYSLSQQVLNFENTLNQYRTMMNG 502
>TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (94%)
Length = 1541
Score = 182 bits (463), Expect = 2e-46
Identities = 117/346 (33%), Positives = 182/346 (51%), Gaps = 16/346 (4%)
Frame = +1
Query: 5 SKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYG 64
S+++L LL V++ + G+ VP IF FGDS+ D GNNN+ T KANF PYG
Sbjct: 373 SRSFLASFLLAVLL------NVTNGQPLVPAIFTFGDSIVDVGNNNHQLTIVKANFPPYG 534
Query: 65 IDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANL--NGSDILKGVNYASGSAGIRK 122
DF FPTGR+ NG+ D IA +LGF + P + NL G ++L G N+AS S+G
Sbjct: 535 RDFENHFPTGRFCNGKLATDFIADILGFTSYQPAYLNLKTKGKNLLNGANFASASSGY-F 711
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFL 182
E S+L ++ L QL +++ +++ G A+ ++ +Y ++ GT+DF QNY++
Sbjct: 712 ELTSKLYSSIPLSKQLEYYKECQTKLV-EAAGQSSASSIISDAIYLISAGTSDFVQNYYI 888
Query: 183 PNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV-----N 237
+ N ++YT Q++ L++ S+++Q+L+ GAR+ + L +GC+P ++
Sbjct: 889 NPLLN--KLYTTDQFSDTLLRCYSNFIQSLYALGARRIGVTSLPPIGCLPAVITLFGAHI 1062
Query: 238 GSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSV-----GFTVTH 292
CV N FN++L + L K +L + + ++D + GF
Sbjct: 1063NECVTSLNSDAINFNEKLNTTSQNL-KNMLPGLNLVVFDIYQPLYDLATKPSENGFFEAR 1239
Query: 293 HGCCPT---NEKGKCIRDGI-PCQNRHEYVFWDGIHTTEAANLVTA 334
CC T C + I C N EYVFWDG H +EAAN V A
Sbjct: 1240KACCGTGLIEVSILCNKKSIGTCANASEYVFWDGFHPSEAANKVLA 1377
>TC217084
Length = 1310
Score = 179 bits (454), Expect = 2e-45
Identities = 119/366 (32%), Positives = 182/366 (49%), Gaps = 15/366 (4%)
Frame = +3
Query: 5 SKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYG 64
S +L L LL+ +++ I+ + FVFGDSL D+GNNN L T+A+A+ PYG
Sbjct: 87 SSKFLCLCLLITLISIIVAPQAEAARA----FFVFGDSLVDNGNNNFLATTARADSYPYG 254
Query: 65 IDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKE 123
ID + +GR+SNG N D I++ +G + +P + LNG +L G N+AS GI +
Sbjct: 255 IDSASHRASGRFSNGLNMPDLISEKIGSEPTLPYLSPQLNGERLLVGANFASAGIGILND 434
Query: 124 SGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLP 183
+G Q + + + QL + + R++ L G ++ +N+ L + +G NDF NY+L
Sbjct: 435 TGIQFINIIRITEQLAYFKQYQQRVS-ALIGEEQTRNLVNKALVLITLGGNDFVNNYYLV 611
Query: 184 NVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLV----NGS 239
SR Y Y LI + L L+ GAR+ ++ G LGC+P L NG
Sbjct: 612 PFSARSREYALPDYVVFLISEYRKILANLYELGARRVLVTGTGPLGCVPAELAMHSQNGE 791
Query: 240 CVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTA-------IIHDKSVGFTVTH 292
C E A LFN QL L+ LN +I D +FI++ A + + ++ GF +
Sbjct: 792 CATELQRAVNLFNPQLVQLLHELNTQIGSD---VFISANAFTMHLDFVSNPQAYGFVTSK 962
Query: 293 HGCC---PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPT 349
CC N G C C NR Y FWD H +E AN + + + + + +P
Sbjct: 963 VACCGQGAYNGIGLCTPASNLCPNRDLYAFWDPFHPSERANRL-IVDKFMTGSTEYMHPM 1139
Query: 350 NIKHLI 355
N+ +I
Sbjct: 1140NLSTII 1157
>TC218564
Length = 1398
Score = 176 bits (445), Expect = 2e-44
Identities = 115/351 (32%), Positives = 188/351 (52%), Gaps = 17/351 (4%)
Frame = +1
Query: 5 SKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYG 64
S WL+L ++VA +T ++ VP + VFGDS DSGNNN + T K+NF PYG
Sbjct: 13 SPAWLLLLTQSLLVAV----TTSEAKNNVPAVIVFGDSSVDSGNNNVIATVLKSNFKPYG 180
Query: 65 IDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSAGIRK 122
DF PTGR+ NGR P D IA+ G + +P + + D GV +AS G
Sbjct: 181 RDFEGDRPTGRFCNGRVPPDFIAEAFGIKRTVPAYLDPAYTIQDFATGVCFASAGTGYDN 360
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFL 182
+ + L + + L ++ +++ +++ L G++KA + +++ LY +++GTNDF +NY+
Sbjct: 361 ATSAVL-NVIPLWKEIEYYKEYQAKLRTHL-GVEKANKIISEALYLMSLGTNDFLENYY- 531
Query: 183 PNVFNTSRM-YTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIP-----KLLV 236
VF T R+ +T QY L++ ++++ L+ G RK + GL +GC+P +L
Sbjct: 532 --VFPTRRLHFTVSQYQDFLLRIAENFVRELYALGVRKLSITGLGPVGCLPLERATNILG 705
Query: 237 NGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVT 291
+ C +E N FN +L++++ +LN++ L K + N+ +I++D + GF V
Sbjct: 706 DHGCNQEYNDVALSFNRKLENVITKLNRE-LPRLKALSANAYSIVNDIITKPSTYGFEVV 882
Query: 292 HHGCCPTN--EKGKCIRDGIP--CQNRHEYVFWDGIHTTEAANLVTAITSY 338
CC T E D P C + +YVFWD H TE N + ++SY
Sbjct: 883 EKACCSTGTFEMSYLCSDKNPLTCTDAEKYVFWDAFHPTEKTNRI--VSSY 1029
>TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase EXL1, partial
(49%)
Length = 1085
Score = 171 bits (434), Expect = 4e-43
Identities = 115/335 (34%), Positives = 177/335 (52%), Gaps = 17/335 (5%)
Frame = +2
Query: 12 SLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDF-PAT 70
S +V+++ + ++ +P + VFGDS+ D+GNNN + T AK NFLPYG DF
Sbjct: 53 SFAIVIISLHVSSVSLPNYESIPAVIVFGDSIVDTGNNNYITTIAKCNFLPYGRDFGGGN 232
Query: 71 FPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQL 128
PTGR+SNG P D IA G +E +PP+ + L D+L GV++ASG++G + S++
Sbjct: 233 QPTGRFSNGLTPSDIIAAKFGVKELLPPYLDPKLQPQDLLTGVSFASGASGYDPLT-SKI 409
Query: 129 GHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNT 188
++L QL R ++I +G AT +++ +Y + G+ND YF+
Sbjct: 410 ASALSLSDQLDTFREYKNKIMEIVGENRTAT-IISKSIYILCTGSNDITNTYFV-----R 571
Query: 189 SRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIP--KLLVNG---SCVEE 243
Y + Y + Q +++LQ L+ GAR+ +VGL LGC+P + L G +C +
Sbjct: 572 GGEYDIQAYTDLMASQATNFLQELYGLGARRIGVVGLPVLGCVPSQRTLHGGIFRACSDF 751
Query: 244 KNVATFLFNDQLKSLVDRLNKKILMDSKFIFIN-----STAIIHDKSVGFTVTHHGCCPT 298
+N A LFN +L S +D L KK +++F++++ I + GF V GCC T
Sbjct: 752 ENEAAVLFNSKLSSQMDAL-KKQFQEARFVYLDLYNPVLNLIQNPAKYGFEVMDQGCCGT 928
Query: 299 N--EKGKCIRDG--IPCQNRHEYVFWDGIHTTEAA 329
E G + C N Y+FWD H TEAA
Sbjct: 929 GKLEVGPLCNHFTLLICSNTSNYIFWDSFHPTEAA 1033
>TC205733
Length = 1377
Score = 169 bits (428), Expect = 2e-42
Identities = 112/346 (32%), Positives = 178/346 (51%), Gaps = 15/346 (4%)
Frame = +2
Query: 4 ESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPY 63
++ T L+ S +VV++ + T ++V + VFGDS D+GNNN +PT A++NF PY
Sbjct: 74 KTTTLLLCSHIVVLLLLSLVAET---SAKVSAVIVFGDSSVDAGNNNFIPTIARSNFQPY 244
Query: 64 GIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSAGIR 121
G DF TGR+ NGR P D I++ G + ++P + + N SD GV +AS + G
Sbjct: 245 GRDFEGGKATGRFCNGRIPTDFISESFGLKPYVPAYLDPKYNISDFASGVTFASAATGYD 424
Query: 122 KESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYF 181
+ L + L QL +++ ++ LG KA + + L+ +++GTNDF +NY+
Sbjct: 425 NATSDVLS-VIPLWKQLEYYKGYQKNLSAYLGE-SKAKDTIAEALHLMSLGTNDFLENYY 598
Query: 182 LPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIP-----KLLV 236
+ + +TP+QY L +++++L+ GARK L GL +GC+P +
Sbjct: 599 --TMPGRASQFTPQQYQNFLAGIAENFIRSLYGLGARKVSLGGLPPMGCLPLERTTSIAG 772
Query: 237 NGSCVEEKNVATFLFNDQLKSLVDRLNKKI----LMDSKFIFINSTAIIHDKSVGFTVTH 292
CV N FN++LK+L +LN+++ L+ S +I + I + GF T
Sbjct: 773 GNDCVARYNNIALEFNNRLKNLTIKLNQELPGLKLVFSNPYYIMLSIIKRPQLYGFESTS 952
Query: 293 HGCCPTN--EKGKCIRDG--IPCQNRHEYVFWDGIHTTEAANLVTA 334
CC T E G G C + +YVFWD H TE N + A
Sbjct: 953 VACCATGMFEMGYACSRGQMFSCTDASKYVFWDSFHPTEMTNSIVA 1090
>TC204290
Length = 823
Score = 167 bits (422), Expect = 9e-42
Identities = 86/206 (41%), Positives = 128/206 (61%), Gaps = 13/206 (6%)
Frame = +2
Query: 169 VNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDRL 228
+ +G+ND+ NYF+P +++SR Y+P +YA LIQ + L+TL+++GARK VL G+ ++
Sbjct: 5 IGLGSNDYLNNYFMPQFYSSSRQYSPDEYADVLIQAYTEQLKTLYNYGARKMVLFGIGQI 184
Query: 229 GCIPKLLVNGS-----CVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD 283
GC P L S CVE+ N A +FN++LK L D+ N + L D+K I+INS I D
Sbjct: 185 GCSPNELAQNSPDGKTCVEKINTANQIFNNKLKGLTDQFNNQ-LPDAKVIYINSYGIFQD 361
Query: 284 -----KSVGFTVTHHGCC---PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAI 335
+ GF+VT+ GCC N + C+ PCQNR EY+FWD H TEA N+V A
Sbjct: 362 IISNPSAYGFSVTNAGCCGVGRNNGQITCLPMQTPCQNRREYLFWDAFHPTEAGNVVVAQ 541
Query: 336 TSYNSSNPAIAYPTNIKHLIQSNTIN 361
+Y++ + + AYP +I+ L Q ++N
Sbjct: 542 RAYSAQSASDAYPVDIQRLAQI*SLN 619
>TC206646
Length = 1178
Score = 155 bits (393), Expect = 2e-38
Identities = 100/303 (33%), Positives = 153/303 (50%), Gaps = 15/303 (4%)
Frame = +1
Query: 45 DSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFAN-LN 103
DSGNN+ L T+A+A+ PYGID+P PTGR+SNG N D I+ LG + +P + L
Sbjct: 7 DSGNNDFLVTTARADAPPYGIDYPTHRPTGRFSNGLNIPDLISLELGLKPTLPYLSPLLV 186
Query: 104 GSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLN 163
G +L G N+AS GI ++G Q + +++ QL R++ +G + N
Sbjct: 187 GEKLLIGANFASAGIGILNDTGIQFLNIIHIQKQLKLFHEYQERLSLHIGA-EGTRNLXN 363
Query: 164 QCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLV 223
+ L + +G NDF NY+L SR ++ Y + LI + L+ L+ GAR+ ++
Sbjct: 364 RALVLITLGGNDFVNNYYLVPYSARSRQFSLPDYVRYLISEYRKVLRRLYDLGARRVLVT 543
Query: 224 GLDRLGCIPKLLV----NGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTA 279
G +GC+P L G C E A LFN QL +++ LN+++ D +FI + A
Sbjct: 544 GTGPMGCVPAELATRSRTGDCDVELQRAASLFNPQLVQMLNGLNQELGAD---VFIAANA 714
Query: 280 -------IIHDKSVGFTVTHHGCC---PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAA 329
+ + ++ GF + CC P N G C C NR Y FWD H +E A
Sbjct: 715 QRMHMDFVSNPRAYGFVTSKIACCGQGPYNGVGLCTPTSNLCPNRDLYAFWDPFHPSEKA 894
Query: 330 NLV 332
+ +
Sbjct: 895 SRI 903
>TC211953 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (37%)
Length = 1023
Score = 149 bits (375), Expect = 3e-36
Identities = 84/231 (36%), Positives = 128/231 (55%), Gaps = 11/231 (4%)
Frame = +1
Query: 137 QLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQ 196
QL + +++I LG D L +C+++V +G+ND+ NY +PN + T Y +Q
Sbjct: 235 QLSNFENTLNQITGNLGA-DYMGTALARCIFFVGMGSNDYLNNYLMPN-YPTRNQYNGQQ 408
Query: 197 YAKALIQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLL---VNGSCVEEKNVATFLFND 253
YA L+Q S L L++ GARK V+ GL +GCIP +L G+C EE N+ FN+
Sbjct: 409 YADLLVQTYSQQLTRLYNLGARKFVIAGLGEMGCIPSILAQSTTGTCSEEVNLLVQPFNE 588
Query: 254 QLKSLVDRLNKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPT-NEKGK--CI 305
+K+++ N L ++FIF +S+ + D +S GF V + GCC +G+ C+
Sbjct: 589 NVKTMLGNFNNN-LPGARFIFADSSRMFQDILLNARSYGFAVVNRGCCGIGRNRGQITCL 765
Query: 306 RDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQ 356
PC NR +YVFWD H TEA N++ ++N NP YP NI+ L +
Sbjct: 766 PFQTPCPNRRQYVFWDAFHPTEAVNILMGRMAFN-GNPNFVYPINIRQLAE 915
>TC207231 similar to UP|Q94CH6 (Q94CH6) Family II lipase EXL3, partial (70%)
Length = 1216
Score = 147 bits (372), Expect = 6e-36
Identities = 95/303 (31%), Positives = 153/303 (50%), Gaps = 15/303 (4%)
Frame = +3
Query: 40 GDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPF 99
GDS+ D GNNN + T K +F PYG DF PTGR+ NG+ P D + + LG +E +P +
Sbjct: 3 GDSIVDPGNNNKVKTLVKCDFPPYGKDFEGGIPTGRFCNGKIPSDLLVEELGIKELLPAY 182
Query: 100 --ANLNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDK 157
NL SD++ GV +ASG++G + ++ +++ QL + + ++ H + G D+
Sbjct: 183 LDPNLKPSDLVTGVCFASGASGYDPLT-PKIASVISMSEQLDMFKEYIGKLKH-IVGEDR 356
Query: 158 ATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGA 217
L + V G++D YF+ V Y Y ++ S++++ L+ GA
Sbjct: 357 TKFILANSFFLVVAGSDDIANTYFIARV--RQLQYDIPAYTDLMLHSASNFVKELYGLGA 530
Query: 218 RKSVLVGLDRLGCIP--KLLVNG---SCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKF 272
R+ ++ +GC+P + L G C EE N A LFN +L +D L K L +S+
Sbjct: 531 RRIGVLSAPPIGCVPSQRTLAGGFQRECAEEYNYAAKLFNSKLSRELDAL-KHNLPNSRI 707
Query: 273 IFIN-----STAIIHDKSVGFTVTHHGCCPTNE---KGKCIRDGIPCQNRHEYVFWDGIH 324
++I+ I++ + G+ V GCC T + C G C + +YVFWD H
Sbjct: 708 VYIDVYNPLMDIIVNYQRHGYKVVDRGCCGTGKLEVAVLCNPLGATCPDASQYVFWDSYH 887
Query: 325 TTE 327
TE
Sbjct: 888 PTE 896
>TC229328
Length = 514
Score = 144 bits (363), Expect = 6e-35
Identities = 69/132 (52%), Positives = 98/132 (73%)
Frame = +2
Query: 14 LVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPT 73
L+V+V S+ V G QVPC F+FGDSL D+GNNN L + A+A++LPYGIDFP P+
Sbjct: 122 LIVVVVSLGLWGGVQGAPQVPCYFIFGDSLVDNGNNNQLQSLARADYLPYGIDFPGG-PS 298
Query: 74 GRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHNVN 133
GR+SNG+ +D IA++LGF ++IPP+A+ +G ILKGVNYAS +AGIR+E+G QLG +
Sbjct: 299 GRFSNGKTTVDAIAELLGFDDYIPPYADASGDAILKGVNYASAAAGIREETGQQLGGRII 478
Query: 134 LGLQLLHHRAIV 145
Q+ ++++ V
Sbjct: 479 FSGQVHNYQSTV 514
>TC228081 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (52%)
Length = 680
Score = 132 bits (331), Expect = 3e-31
Identities = 76/216 (35%), Positives = 127/216 (58%), Gaps = 2/216 (0%)
Frame = +3
Query: 5 SKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYG 64
S ++ SLL+V+V ++ + G+ VP +F+FGDS+ D GNNN+L T KANF PYG
Sbjct: 60 SSSYFFTSLLLVVVFNLAK-----GQPLVPALFIFGDSVVDVGNNNHLYTIVKANFPPYG 224
Query: 65 IDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANL--NGSDILKGVNYASGSAGIRK 122
DF PTGR+ NG+ D A+ LGF + P + NL G+++L G N+AS ++G
Sbjct: 225 RDFKNHNPTGRFCNGKLASDYTAENLGFTSYPPAYLNLKAKGNNLLNGANFASAASGY-Y 401
Query: 123 ESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFL 182
+ ++L H + L QL H++ + + + G A+ ++ +Y ++ G +DF QNY++
Sbjct: 402 DPTAKLYHAIPLSQQLEHYKECQNILVGTV-GQPNASSIISGAIYLISAGNSDFIQNYYI 578
Query: 183 PNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGAR 218
+ ++YT Q++ L+Q + ++Q L+ GAR
Sbjct: 579 NPLL--YKVYTADQFSDILLQSYATFIQNLYALGAR 680
>TC225398 similar to UP|O04320 (O04320) Proline-rich protein APG isolog,
partial (93%)
Length = 1118
Score = 113 bits (283), Expect(2) = 8e-31
Identities = 77/262 (29%), Positives = 128/262 (48%), Gaps = 17/262 (6%)
Frame = +3
Query: 90 LGFQEFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSR 147
+GFQ + P + + +G ++L G N+AS ++G E+ + L H + L QL + + +
Sbjct: 114 IGFQTYAPAYLSPQASGKNLLIGANFASAASGY-DENAATLNHAIPLSQQLSYFKEYQGK 290
Query: 148 IAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSH 207
+A ++ G KA + LY ++ G++DF QNY++ N ++Y+P QY+ L+ + S
Sbjct: 291 LA-KVAGSKKAASIIKDALYVLSAGSSDFVQNYYVNPWIN--KVYSPDQYSSYLVGEFSS 461
Query: 208 YLQTLHHFGARKSVLVGLDRLGCIPKLLV-----NGSCVEEKNVATFLFNDQLKSLVDRL 262
+++ L+ GAR+ + L LGC+P CV N FN +L S L
Sbjct: 462 FVKDLYGLGARRLGVTSLPPLGCLPAARTIFGFHENGCVSRINTDAQGFNKKLNSAAASL 641
Query: 263 NKKILMDSKFIFINSTAIIHD-----KSVGFTVTHHGCCPTN--EKGKCI---RDGIPCQ 312
K+ L K + ++D GF + GCC T E + + C
Sbjct: 642 QKQ-LPGLKIAIFDIYKPLYDLVQSPSKSGFVEANRGCCGTGTVETTSLLCNSKSPGTCS 818
Query: 313 NRHEYVFWDGIHTTEAANLVTA 334
N +YVFWD +H ++AAN V A
Sbjct: 819 NATQYVFWDSVHPSQAANQVLA 884
Score = 38.1 bits (87), Expect(2) = 8e-31
Identities = 18/40 (45%), Positives = 24/40 (60%)
Frame = +1
Query: 57 KANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFI 96
KA++ PYG DF PTGR+ NG+ D A LGF+ +
Sbjct: 16 KADYPPYGRDFVNHQPTGRFCNGKLATDFTADTLGFRPML 135
>TC226732
Length = 985
Score = 129 bits (325), Expect = 2e-30
Identities = 88/266 (33%), Positives = 128/266 (48%), Gaps = 14/266 (5%)
Frame = +3
Query: 108 LKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLY 167
L G N+AS GI ++G Q + + + QL + + +R++ +G +A + Q L
Sbjct: 9 LVGANFASAGIGILNDTGVQFVNVIRMYRQLEYFKEYQNRVSAIIGA-SEAKNLVKQALV 185
Query: 168 YVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLVGLDR 227
+ +G NDF NYFL S+ Y Y K LI + LQ L+ GAR+ ++ G
Sbjct: 186 LITVGGNDFVNNYFLVPNSARSQQYPLPAYVKYLISEYQKLLQRLYDLGARRVLVTGTGP 365
Query: 228 LGCIPKLLV----NGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHD 283
LGC+P L NG CV E A LFN QL+ ++ +LN+KI D FI N+ +D
Sbjct: 366 LGCVPSELAQRGRNGQCVPELQQAAALFNPQLEQMLLQLNRKIATD-VFIAANTGKAHND 542
Query: 284 -----KSVGFTVTHHGCC---PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAI 335
+ GF + CC P N G C C NR +Y FWD H +E AN + +
Sbjct: 543 FVTNAQQFGFVTSQVACCGQGPYNGIGLCTALSNLCSNRDQYAFWDAFHPSEKANRL-IV 719
Query: 336 TSYNSSNPAIAYPTNIKHL--IQSNT 359
S + A P N+ + + SNT
Sbjct: 720 EEIMSGSKAYMNPMNLSTILALDSNT 797
>TC207585 similar to UP|Q9AYM7 (Q9AYM7) CPRD47 protein, partial (73%)
Length = 840
Score = 129 bits (324), Expect = 2e-30
Identities = 92/281 (32%), Positives = 142/281 (49%), Gaps = 16/281 (5%)
Frame = +1
Query: 32 QVPCIFVFGDSLSDSGNNNNLPTSAKANFLP-YGIDFPATFPTGRYSNGRNPIDKIAQML 90
+ P ++VFGDSL D GNNN L S + LP YGIDFP PTGR+SNG+N D IA+ L
Sbjct: 4 KAPAVYVFGDSLVDVGNNNYLSLSIEKAILPHYGIDFPTKKPTGRFSNGKNAADLIAENL 183
Query: 91 GFQEFIPPFANL---------NGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHH 141
G PP+ +L L GVN+ASG AGI S ++ L Q+ ++
Sbjct: 184 GLPT-SPPYLSLVSKVHNNNKKNVSFLGGVNFASGGAGIFNASDKGFRQSIPLPKQVDYY 360
Query: 142 RAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKAL 201
+ ++ ++G ++L++ ++ V IG ND YF + + + TP+QY ++
Sbjct: 361 SQVHEQLIQQIGA-STLGKHLSKSIFIVVIGGNDI-FGYF--DSKDLQKKNTPQQYVDSM 528
Query: 202 IQQLSHYLQTLHHFGARKSVLVGLDRLGCIPKLLVNG--SCVEEKNVATFLFNDQLKSLV 259
L LQ L++ GA+K + G+ +GC P V CV E N + +N+ L+S++
Sbjct: 529 ASTLKVQLQRLYNNGAKKFEIAGVGAIGCCPAYRVKNKTECVSEANDLSVKYNEALQSML 708
Query: 260 D--RLNKKILMDSKF-IFINSTAIIHD-KSVGFTVTHHGCC 296
RL K + S F + ++H+ S + CC
Sbjct: 709 KEWRLENKHISYSYFDTYAGLQDLVHNPASYRYAYEKAACC 831
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,211,011
Number of Sequences: 63676
Number of extensions: 245836
Number of successful extensions: 1397
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 1249
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1257
length of query: 361
length of database: 12,639,632
effective HSP length: 98
effective length of query: 263
effective length of database: 6,399,384
effective search space: 1683037992
effective search space used: 1683037992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0476a.9


