

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.5
(268 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
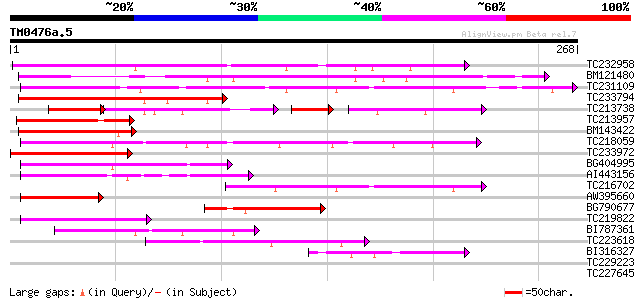
Score E
Sequences producing significant alignments: (bits) Value
TC232958 161 3e-40
BM121480 129 1e-30
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 106 1e-23
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 99 2e-21
TC213738 45 1e-18
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 86 2e-17
BM143422 80 7e-16
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 78 5e-15
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 69 2e-12
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 62 2e-10
AI443156 62 4e-10
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 60 1e-09
AW395660 60 1e-09
BG790677 50 1e-06
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 47 9e-06
BI787361 46 2e-05
TC223618 44 8e-05
BI316327 44 1e-04
TC229223 similar to UP|Q9XI00 (Q9XI00) F8K7.20 protein (At1g2176... 39 0.002
TC227645 similar to UP|Q9FZK1 (Q9FZK1) F17L21.13, partial (61%) 34 0.060
>TC232958
Length = 758
Score = 161 bits (407), Expect = 3e-40
Identities = 103/239 (43%), Positives = 137/239 (57%), Gaps = 23/239 (9%)
Frame = +1
Query: 2 EKLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLL--- 58
+K+ LP +LI EILLRLPV+SL+RFK VCKSW +LISDP+FAKSHFDL AA R+L
Sbjct: 49 KKMVLPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIA 228
Query: 59 -----LRCLFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVW 113
LR + SL D+SA V + + P + + +GSCRGF+LL + + VW
Sbjct: 229 SSAPELRSIDFNASLHDDSASVAVTVDLPAPKPYFHFVEIIGSCRGFILL-HCLSHLCVW 405
Query: 114 NPSTGFHKQILTSCDF------IIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHA----- 162
NP+TG HK + S F L GFGYD STDD+ L+V A N K A
Sbjct: 406 NPTTGVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDDF---LVVHACYNPKHQANCAEI 576
Query: 163 YSVKTNSW-DYNHVHVLYMHLGCDYRH---GVFLNNSLHWLVISKVTSLQVVIAYDLSE 217
+S++ N+W +H Y H R+ G FLN ++HWL S+ V++A+DL E
Sbjct: 577 FSLRANAWKGIEGIHFPYTHFRYTNRYNQFGSFLNGAIHWLAFRINASINVIVAFDLVE 753
>BM121480
Length = 917
Score = 129 bits (324), Expect = 1e-30
Identities = 97/263 (36%), Positives = 128/263 (47%), Gaps = 12/263 (4%)
Frame = +3
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFD 64
TLP ELI EILLRLPV+SLLRFKCV L C D
Sbjct: 63 TLPQELIREILLRLPVKSLLRFKCV---------------------------LFKTCSRD 161
Query: 65 LDSLGDNSAVVTLNLPPPCMSCIQNSLY-----FLGSCRGFMLLA-NEYQRVIVWNPSTG 118
VV LP P + C++ + LGSCRG +LL ++ +I+WNPS G
Sbjct: 162 ---------VVYFPLPLPSIPCLRLDDFGIRPKILGSCRGLVLLYYDDSANLILWNPSLG 314
Query: 119 FHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHA--YSVKTNSWDYNHV- 175
HK + D I YGFGYD S D+Y L+LI L + A YS KT SW + +
Sbjct: 315 RHKXLPNYRDDITSFPYGFGYDESKDEYLLILIGLPKFGPETGADIYSFKTESWKTDTIV 494
Query: 176 -HVLYMHLGCDY--RHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLLPELAEP 232
L + D R G LN +LHW V S+ V+IA+DL E +LS IP LP
Sbjct: 495 YDPLXRYXAEDXIARAGSLLNGALHWFVFSESKXDHVIIAFDLVERTLSXIP-LPLADRS 671
Query: 233 VLKAECMPTFYQVRVLGGCLSLC 255
++ + + Y ++++G CL +C
Sbjct: 672 TVQKDAV---YGLKIMGXCLXVC 731
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 106 bits (265), Expect = 1e-23
Identities = 83/282 (29%), Positives = 142/282 (49%), Gaps = 19/282 (6%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLR---CL 62
LP E++ EIL RLPV+S++R + CK W +I F H + + + L+LR L
Sbjct: 93 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKSHS---SLILRHRSHL 263
Query: 63 FDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQ 122
+ LD V L+ P + C NS+ LGS G + ++N + +WNP H+
Sbjct: 264 YSLDLKSPEQNPVELSHP---LMCYSNSIKVLGSSNGLLCISNVADDIALWNPFLRKHR- 431
Query: 123 ILTSCDF-------IIGSLYGFGYDNSTDDYFLVLIVL------ASMNAKIHAYSVKTNS 169
IL + F +YGFG+ + ++DY L+ I + ++++ Y++K++S
Sbjct: 432 ILPADRFHRPQSSLFAARVYGFGHHSPSNDYKLLSITYFVDLQKRTFDSQVQLYTLKSDS 611
Query: 170 WDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQ--VVIAYDLSETSLSEIPLLP 227
W ++ + L C GVF++ SLHWLV K+ + +++++DL+ + E+PL
Sbjct: 612 W--KNLPSMPYALCCARTMGVFVSGSLHWLVTRKLQPHEPDLIVSFDLTRETFHEVPL-- 779
Query: 228 ELAEPVLKAECMPTFYQVRVLGGCLSLC-YTGSRWDMGHERI 268
PV QV +LGGCL + + G+ +D+ R+
Sbjct: 780 ----PVTVNGDFD--MQVALLGGCLCVVEHRGTGFDVWVMRV 887
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 99.4 bits (246), Expect = 2e-21
Identities = 60/112 (53%), Positives = 71/112 (62%), Gaps = 13/112 (11%)
Frame = +1
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCL-F 63
TLP ELI EILLRLPV+SLLRFKCVCKS+ LISDPQF SH+ L A+PTHRL+LR F
Sbjct: 106 TLPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAASPTHRLILRSHDF 285
Query: 64 DLDSLGDNSA-------VVTLNLPPPCMSCIQNSLY-----FLGSCRGFMLL 103
S+ S VV LP P + C++ + LGSCRG +LL
Sbjct: 286 YAQSIATESVFKTCSRDVVYFPLPLPSIPCLRLDDFGIRPKILGSCRGLVLL 441
>TC213738
Length = 690
Score = 45.4 bits (106), Expect(4) = 1e-18
Identities = 21/27 (77%), Positives = 21/27 (77%)
Frame = -1
Query: 19 PVRSLLRFKCVCKSWFYLISDPQFAKS 45
PVR LLR K VCKSW LI DPQFAKS
Sbjct: 663 PVRCLLRLKYVCKSWLSLIPDPQFAKS 583
Score = 44.3 bits (103), Expect(4) = 1e-18
Identities = 33/92 (35%), Positives = 46/92 (49%), Gaps = 9/92 (9%)
Frame = -3
Query: 45 SHFDLNAAPTHRLLLRCL----FDLDS--LGDNSAVVTLNLP---PPCMSCIQNSLYFLG 95
S+FDL PTH+LLL+ + F+ +S + D+S V N+P P + +G
Sbjct: 583 SNFDLAVTPTHKLLLKDISNSYFEANSVDIDDDSTDVKFNIPNASPTFKYEFWVGVVVVG 404
Query: 96 SCRGFMLLANEYQRVIVWNPSTGFHKQILTSC 127
SCRGF+LL NP T K IL+ C
Sbjct: 403 SCRGFVLLK---------NPHTSHSKNILSLC 335
Score = 30.8 bits (68), Expect(4) = 1e-18
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 9/74 (12%)
Frame = -2
Query: 161 HAYSVKTNSWDY--NHVHVLYMHLGCDYRHGVFLNNS-------LHWLVISKVTSLQVVI 211
+ +S ++NSW Y N + M R G+ N LHWL K ++I
Sbjct: 245 YCFSWRSNSWSYIKNK*RMFSMISPI*KRGGMIGFNMGHY*MGVLHWLASLKREY*GIII 66
Query: 212 AYDLSETSLSEIPL 225
A+++++ SL EIPL
Sbjct: 65 AFNVTQRSLLEIPL 24
Score = 28.9 bits (63), Expect(4) = 1e-18
Identities = 11/20 (55%), Positives = 15/20 (75%)
Frame = -1
Query: 134 LYGFGYDNSTDDYFLVLIVL 153
LYG GYD+S +DY +V + L
Sbjct: 309 LYGIGYDSSMNDYMVVAVSL 250
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 85.9 bits (211), Expect = 2e-17
Identities = 43/56 (76%), Positives = 48/56 (84%)
Frame = +3
Query: 4 LTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLL 59
+TLP ELI EILLR PVRS+LRFKCVCKSW LISDPQF +HFDL A+PTHRL+L
Sbjct: 27 VTLPLELIREILLRSPVRSVLRFKCVCKSWLSLISDPQF--THFDLAASPTHRLIL 188
Score = 38.1 bits (87), Expect = 0.004
Identities = 23/66 (34%), Positives = 32/66 (47%), Gaps = 7/66 (10%)
Frame = +1
Query: 135 YGFGYDNSTDDYFLVLIV-------LASMNAKIHAYSVKTNSWDYNHVHVLYMHLGCDYR 187
YG YD TDD L++ L +I +S KTN +++V Y HLG ++
Sbjct: 289 YGLVYDELTDD*STCLLLFQLFSDSLDECKPEIQVFSFKTNLRSRYNINVPYKHLGLEFG 468
Query: 188 HGVFLN 193
G FLN
Sbjct: 469 AGSFLN 486
>BM143422
Length = 424
Score = 80.5 bits (197), Expect = 7e-16
Identities = 41/57 (71%), Positives = 46/57 (79%), Gaps = 1/57 (1%)
Frame = +1
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLN-AAPTHRLLLR 60
TLP +LI ILL+LPV+S+ RFKCVCKSW LISDPQF SHFDL A P+HRLLLR
Sbjct: 226 TLPLDLIELILLKLPVKSVTRFKCVCKSWLSLISDPQFGFSHFDLALAVPSHRLLLR 396
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 77.8 bits (190), Expect = 5e-15
Identities = 74/257 (28%), Positives = 113/257 (43%), Gaps = 39/257 (15%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHF----DLNAAPTHRLLLRC 61
LP EL+ +L RLP + LL KCVCKSWF LI+DP F +++ L + H L++R
Sbjct: 66 LPGELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHLLVIRR 245
Query: 62 LFDLDSLGDNSAVVTLNLPPP---CMSCIQNSLY-----------FLGSCRGFMLLANEY 107
F L +V++ N P S + N Y LG C G L E
Sbjct: 246 PF-FSGLKTYISVLSWNTNDPKKHVSSDVLNPPYEYNSDHKYWTEILGPCNGIYFL--EG 416
Query: 108 QRVIVWNPSTGFHKQILTS------CDFIIGSLYGFGYDNSTDDYFLVLI---------- 151
++ NPS G K + S + GFG+D T+DY +V++
Sbjct: 417 NPNVLMNPSLGEFKALPKSHFTSPHGTYTFTDYAGFGFDPKTNDYKVVVLKDLWLKETDE 596
Query: 152 -VLASMNAKIHAYSVKTNSWDYNHVHVLYM--HLGCDYRHGVFLNNSLHW--LVISKVTS 206
+ +A++ YS+ +NSW +L + + R + NN HW V +
Sbjct: 597 REIGYWSAEL--YSLNSNSWRKLDPSLLPLPIEIWGSSRVFTYANNCCHWWGFVEESDAT 770
Query: 207 LQVVIAYDLSETSLSEI 223
VV+A+D+ + S +I
Sbjct: 771 QDVVLAFDMVKESFRKI 821
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 68.9 bits (167), Expect = 2e-12
Identities = 37/58 (63%), Positives = 42/58 (71%)
Frame = -2
Query: 1 MEKLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLL 58
M L LP E+I EILLRLPV+SL FK VCKS LIS+P FAK HF+ NAA T +LL
Sbjct: 555 MVXLFLPQEIIIEILLRLPVKSLPSFKFVCKS*LSLISNPHFAKWHFERNAAHTEKLL 382
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 62.4 bits (150), Expect = 2e-10
Identities = 41/102 (40%), Positives = 52/102 (50%), Gaps = 2/102 (1%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHF--DLNAAPTHRLLLRCLF 63
LP E++ EIL RLPVRSLLRF+ KSW LI H L A L+LR
Sbjct: 40 LPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLASNTSLILRVDS 219
Query: 64 DLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLAN 105
DL + ++L P M C NS+ LGSC G + ++N
Sbjct: 220 DLYQTNFPTLDPPVSLNHPLM-CYSNSITLLGSCNGLLCISN 342
>AI443156
Length = 414
Score = 61.6 bits (148), Expect = 4e-10
Identities = 44/115 (38%), Positives = 59/115 (51%), Gaps = 5/115 (4%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPT-----HRLLLR 60
LP E++ EIL RLPV+S++R + CK W +I F H LN + T HR L
Sbjct: 90 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFH--LNKSHTSLILRHRSQLY 263
Query: 61 CLFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNP 115
L DL SL D + L P M C NS+ LGS G + ++N + +WNP
Sbjct: 264 SL-DLKSLLDPN---PFELSHPLM-CYSNSIKVLGSSNGLLCISNVXDDIALWNP 413
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 60.1 bits (144), Expect = 1e-09
Identities = 42/140 (30%), Positives = 66/140 (47%), Gaps = 17/140 (12%)
Frame = +3
Query: 103 LANEYQRVIVWNPSTGFHKQIL---------TSCDFIIGSLYGFGYDNSTDDYFLVLIVL 153
++N + WNPS H+ + + GFG+D+ T DY LV I
Sbjct: 9 ISNVADDIAFWNPSLRQHRILPYLPVPRRRHPDTTLFAARVCGFGFDHKTRDYKLVRISY 188
Query: 154 ------ASMNAKIHAYSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSL 207
S +A++ Y+++ N+W + L L C GVF+ NSLHW+V K+
Sbjct: 189 FVDLHDRSFDAQVKLYTLRANAW--KTLPSLPYALCCARTMGVFVGNSLHWVVTRKLEPD 362
Query: 208 Q--VVIAYDLSETSLSEIPL 225
Q ++IA+DL+ E+PL
Sbjct: 363 QPDLIIAFDLTHDIFRELPL 422
>AW395660
Length = 381
Score = 59.7 bits (143), Expect = 1e-09
Identities = 27/39 (69%), Positives = 31/39 (79%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAK 44
LP EL+ EIL RLPV+SLL+F+CVCKSW LI DP F K
Sbjct: 264 LPDELVVEILSRLPVKSLLQFRCVCKSWMSLIYDPYFMK 380
>BG790677
Length = 419
Score = 50.1 bits (118), Expect = 1e-06
Identities = 30/58 (51%), Positives = 36/58 (61%), Gaps = 1/58 (1%)
Frame = +1
Query: 93 FLGSCRGFMLLANEYQRV-IVWNPSTGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLV 149
F+ SCR F+LL +Y R ++NPSTG HK + S YGFGYD STDDY LV
Sbjct: 187 FVCSCRDFLLL--DYNRSSFIFNPSTGLHKFVDWSPISNNMGGYGFGYDPSTDDYLLV 354
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 47.0 bits (110), Expect = 9e-06
Identities = 22/62 (35%), Positives = 35/62 (55%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP L+ EIL+ + V + L+ +CVCK W L+ DPQF K H + + L + D+
Sbjct: 196 LPEGLMIEILVWIRVSNPLQLRCVCKRWKSLVVDPQFVKKHLHTSLSDITDLASNAMEDM 375
Query: 66 DS 67
++
Sbjct: 376 NA 381
>BI787361
Length = 423
Score = 45.8 bits (107), Expect = 2e-05
Identities = 31/116 (26%), Positives = 54/116 (45%), Gaps = 19/116 (16%)
Frame = +3
Query: 22 SLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLL-LRCLFDLDSLGDNSAVVTLNLP 80
+L+RF+ V ++W LI DP F K H + + TH LL + ++D D +G V ++
Sbjct: 3 ALMRFRYVSETWNSLIFDPTFVKLHLERSPKNTHVLLEFQAIYDRD-VGQQVGVAPCSIR 179
Query: 81 ----------PPCMSCIQNSLYFLGSCRGFMLLA--------NEYQRVIVWNPSTG 118
C++ +++ GSC G + + E + +WNP+TG
Sbjct: 180 RLVENPSFTIDDCLTLFKHTNSIFGSCNGLVCMTKCFDVREFEEECQYRLWNPATG 347
>TC223618
Length = 444
Score = 43.9 bits (102), Expect = 8e-05
Identities = 32/113 (28%), Positives = 54/113 (47%), Gaps = 7/113 (6%)
Frame = +1
Query: 65 LDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQ-- 122
L SL +N + V L P + ++ +GSC G + A + V++WNPS K+
Sbjct: 46 LSSLFNNLSTVCDELNYPVKNKFRHD-GIVGSCNGLLCFAIKGDCVLLWNPSIRVSKKSP 222
Query: 123 -ILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLAS----MNAKIHAYSVKTNSW 170
+ + + +G GYD+ +DY +V + + K+ YS+ TNSW
Sbjct: 223 PLGNNWRPGCFTAFGLGYDHVNEDYKVVAVFCDPSEYFIECKVKVYSMATNSW 381
>BI316327
Length = 410
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/91 (31%), Positives = 46/91 (49%), Gaps = 15/91 (16%)
Frame = +1
Query: 142 STDDYFLVLIVLASMNAKI---------HAYSVKTNSWD------YNHVHVLYMHLGCDY 186
+TDDY L+VLAS N +S++ N+W +++ Y + D
Sbjct: 94 TTDDY---LVVLASYNRNFPQDELVTHFEYFSLRANTWKATDGTGFSYKQCYYYN---DN 255
Query: 187 RHGVFLNNSLHWLVISKVTSLQVVIAYDLSE 217
+ G F NN+LHWL SL V++A+DL++
Sbjct: 256 QIGCFSNNALHWLAFRFDESLNVIVAFDLTK 348
>TC229223 similar to UP|Q9XI00 (Q9XI00) F8K7.20 protein (At1g21760), partial
(96%)
Length = 1096
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/38 (42%), Positives = 23/38 (60%)
Frame = +1
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQF 42
+LP EL+FE+ R+ L + CVC+ W Y I +P F
Sbjct: 319 SLPDELLFEVFARMTPYDLGKASCVCRKWKYTIRNPVF 432
>TC227645 similar to UP|Q9FZK1 (Q9FZK1) F17L21.13, partial (61%)
Length = 1671
Score = 34.3 bits (77), Expect = 0.060
Identities = 11/38 (28%), Positives = 23/38 (59%)
Frame = +3
Query: 7 PHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAK 44
P +L ++ RLP+ + RF+ VC+ W +++ F++
Sbjct: 366 PEDLFEAVIARLPISTFFRFRSVCRQWNSMLNSQSFSQ 479
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,929,370
Number of Sequences: 63676
Number of extensions: 276097
Number of successful extensions: 1896
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 1868
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1873
length of query: 268
length of database: 12,639,632
effective HSP length: 96
effective length of query: 172
effective length of database: 6,526,736
effective search space: 1122598592
effective search space used: 1122598592
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0476a.5


