

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0437b.7
(824 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
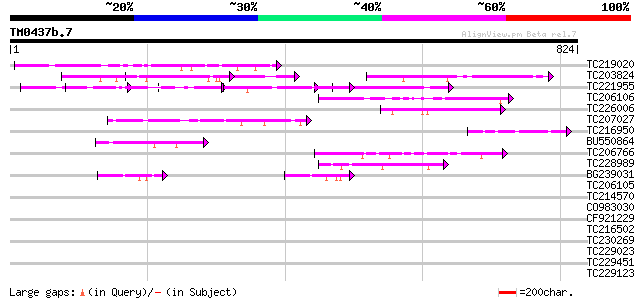
Score E
Sequences producing significant alignments: (bits) Value
TC219020 similar to UP|SRB7_SCHPO (O94376) RNA polymerase II med... 69 1e-11
TC203824 similar to UP|Q9V3T8 (Q9V3T8) SR family splicing factor... 57 3e-08
TC221955 weakly similar to UP|Q6UAL1 (Q6UAL1) Myosin heavy chain... 55 2e-07
TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), ... 52 1e-06
TC226006 similar to UP|REMO_SOLTU (P93788) Remorin (pp34), parti... 49 1e-05
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 47 4e-05
TC216950 46 8e-05
BU550864 45 1e-04
TC206766 similar to PIR|T47576|T47576 FKBP12 interacting protein... 45 1e-04
TC228989 similar to UP|Q9LSS5 (Q9LSS5) Myosin heavy chain-like, ... 45 2e-04
BG239031 44 3e-04
TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-in... 42 0.001
TC214570 homologue to PIR|T05766|T05766 peptidylprolyl isomerase... 41 0.002
CO983030 40 0.004
CF921229 39 0.012
TC216502 similar to GB|AAF40452.1|7211981|F13M7 ESTs gb|N65605, ... 38 0.016
TC230269 weakly similar to UP|WSC2_YEAST (P53832) Cell wall inte... 38 0.021
TC229023 37 0.027
TC229451 similar to UP|Q9FH68 (Q9FH68) Arabidopsis thaliana geno... 37 0.027
TC229123 37 0.046
>TC219020 similar to UP|SRB7_SCHPO (O94376) RNA polymerase II mediator
complex protein srb7, partial (14%)
Length = 1446
Score = 68.6 bits (166), Expect = 1e-11
Identities = 102/434 (23%), Positives = 184/434 (41%), Gaps = 46/434 (10%)
Frame = +2
Query: 8 IPEPANCCSKCEELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQR 67
+ + +N S+ E ++ + + A + K LE + + E K L+
Sbjct: 98 LDDVSNLTSELEAIQARASTLETTLQAANERGKELEDSLNAVTEEKKNLE--------DA 253
Query: 68 KVAIEGKL-EISNTFAALENEVSALISENKKLKQDILE-EQAQGKICDQLKKCEK--VVE 123
+++ KL E N L ++++ + + + D+ E E + +I ++LK E+ VV
Sbjct: 254 SISLNEKLAEKENLLEILRDDLNLTQDKLQSTESDLREAELRESEIIEKLKASEENLVVR 433
Query: 124 GRNALRQAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSA 183
GR+ A + SE + L + + + Q+ ++ +EK N N+ S
Sbjct: 434 GRDIEETATR---------HSELQLLHESLTRDSEQK---LQEAIEKFN------NKDSE 559
Query: 184 LKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQ 243
++S +K+K ILEEQ + A E + N F +++++L+SEN+ LK+ ILD +++
Sbjct: 560 VQSLLEKIK--ILEEQIAK--AGEQSTSLKNEFEESLSKLTSLESENEDLKRQILDAESK 727
Query: 244 G--KFCD---------RLKKKCEKVVEGRN-------ALRQAVKILEKGIENLESENKKL 285
F + +LK K +++ E N A Q + + I L K
Sbjct: 728 SSQSFSENELLVGTNIQLKTKIDELEESLNHALSEKEAAAQELVSHKNSITELNDLQSK- 904
Query: 286 KKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKK-----DILEEQAQRKV 340
+IQ + R +++E +L+ A + S K N+KL + EE A+ V
Sbjct: 905 SSEIQRANEARTLKVESQLQ--EALQRHTEKESETKELNEKLNTLEGQIKLFEEHAREAV 1078
Query: 341 AMEG--KLEISNAFAAL-----------------ENEVSALKSENKKLKQDILEEQAQGK 381
A G K E+ + L E E + L EN KL Q+I ++ K
Sbjct: 1079ATSGTHKAELEQSLIKLKHLEIVIEELQNKSLHHEKETAGLNEENSKLNQEIASYES--K 1252
Query: 382 FCDQLKKKCEKVVE 395
D +K +VE
Sbjct: 1253LSDLQEKLSAALVE 1294
>TC203824 similar to UP|Q9V3T8 (Q9V3T8) SR family splicing factor SC35
(CG5442-PB) (LD32469p), partial (6%)
Length = 1532
Score = 57.0 bits (136), Expect = 3e-08
Identities = 63/269 (23%), Positives = 121/269 (44%), Gaps = 18/269 (6%)
Frame = +2
Query: 76 EISNTFAALENEVSALISENKKLKQDI--LEEQAQGKICDQLKKCEKVVEGRNALRQ--- 130
E + AAL+ E L+SEN K++I L + G D + EK+ E + + Q
Sbjct: 278 ESATKIAALQRERDELVSENAARKEEIKKLTAELDGLRSDGADRSEKIEELQREVAQSRD 457
Query: 131 ---AVKILEKRIENLESENKKLKKDI------QEEQAQRKIEIEGKL-EKSNAFAALENE 180
A +++ R +LE+E +L D+ EE E+ L E + +LENE
Sbjct: 458 AAKATEVIAARAADLETEVARLHHDMVAEMTAAEEARADAAELRKVLGETESRVESLENE 637
Query: 181 VSALKSENKKLKKDILEEQAQRKVAM--EGKLEISNAFAALENEV-SALKSENKKLKQDI 237
++ LK K + D+ +RK+ + ++E N E E + + +++K
Sbjct: 638 LAGLK--KVKAESDVRVRDLERKIGVLETKEIEERNKRIRFEEETRDKIDEKEREIKGYR 811
Query: 238 LDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRK 297
+ K K + +++V+ + L +A++ E+ + LES +L+K+ +E +
Sbjct: 812 QKVEELEKVAAEKKTELDELVKQKLRLEEALRESEEKVTGLESSILQLRKEAKE---AER 982
Query: 298 VEIEGKLEISNAFAALENEVSALKSENKK 326
V I + +++ ++ + SE KK
Sbjct: 983 VIISLNEKAVETVESIDRGLNGVHSEGKK 1069
Score = 50.8 bits (120), Expect = 2e-06
Identities = 58/283 (20%), Positives = 127/283 (44%), Gaps = 11/283 (3%)
Frame = +2
Query: 519 DVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEARKL----LEAA 574
D V G+ D E + K +++E+ SE +E KK AE L + +
Sbjct: 233 DEGVANGGVDDQYGEESATKIAALQRERDELVSENAARKEEIKKLTAELDGLRSDGADRS 412
Query: 575 KKIAPEKAVIPEPANCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKK 634
+KI + + + + + + + + L + E ++ +L+K
Sbjct: 413 EKIEELQREVAQSRDAAKATEVIAARAADLETEVARLHHDMVAEMTAAEEARADAAELRK 592
Query: 635 -----ENEVSALKSEISALQQKCGAGAREGNGDVEVLKAGISDTKKEVNRLKKEHVEE-- 687
E+ V +L++E++ L++ + DV V D ++++ L+ + +EE
Sbjct: 593 VLGETESRVESLENELAGLKKV------KAESDVRV-----RDLERKIGVLETKEIEERN 739
Query: 688 ERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEKQVNERKM 747
+RI + E + +DE++ R+ +E +K+AAE + ++ ++ +++ LE+ + E +
Sbjct: 740 KRIRFEEETRDKIDEKEREIKGYRQKVEELEKVAAEKKTELDELVKQKLRLEEALRESEE 919
Query: 748 KLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQERSES 790
K+ S + K EA++ + ++ +N +KA E ES
Sbjct: 920 KVTGLESSILQLRK--EAKEAERVIISLN-----EKAVETVES 1027
Score = 50.1 bits (118), Expect = 4e-06
Identities = 63/290 (21%), Positives = 120/290 (40%), Gaps = 38/290 (13%)
Frame = +2
Query: 169 EKSNAFAALENEVSALKSENKKLKKDILE-------------------EQAQRKVAMEGK 209
E + AAL+ E L SEN K++I + E+ QR+VA
Sbjct: 278 ESATKIAALQRERDELVSENAARKEEIKKLTAELDGLRSDGADRSEKIEELQREVAQ--S 451
Query: 210 LEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGRNALRQAVK 269
+ + A + + L++E +L D++ E E+ LR+ +
Sbjct: 452 RDAAKATEVIAARAADLETEVARLHHDMVAEMT----------AAEEARADAAELRKVLG 601
Query: 270 ILEKGIENLESENKKLKK-----DIQEEHAQRKV------EIEGK-------LEISNAFA 311
E +E+LE+E LKK D++ +RK+ EIE + E +
Sbjct: 602 ETESRVESLENELAGLKKVKAESDVRVRDLERKIGVLETKEIEERNKRIRFEEETRDKID 781
Query: 312 ALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQ 371
E E+ + + ++L+K E++ + ++ KL + A E +V+ L+S +L++
Sbjct: 782 EKEREIKGYRQKVEELEKVAAEKKTELDELVKQKLRLEEALRESEEKVTGLESSILQLRK 961
Query: 372 DILEEQAQGKFCDQLKKKCEKVVEGRNALR-QAVKILEKGIENLESENKK 420
+ K+ E+V+ N + V+ +++G+ + SE KK
Sbjct: 962 E--------------AKEAERVIISLNEKAVETVESIDRGLNGVHSEGKK 1069
>TC221955 weakly similar to UP|Q6UAL1 (Q6UAL1) Myosin heavy chain class XI E3
protein, partial (6%)
Length = 728
Score = 54.7 bits (130), Expect = 2e-07
Identities = 43/163 (26%), Positives = 75/163 (45%)
Frame = +1
Query: 16 SKCEELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEGKL 75
+K E L K E++ L++ +K E+ +E+EN+ K+ +E Q +
Sbjct: 154 TKLELLTNKNEELETEVEELKKKIKEFEESYSEIENENQARLKEAEEAQLKAT------- 312
Query: 76 EISNTFAALENEVSALISENKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQAVKIL 135
++ T LE +S L SEN+ L Q LEE ++ ++ +KIL
Sbjct: 313 QLQETIERLELSLSNLESENQVLCQKALEEPKNEELFEE-----------------IKIL 441
Query: 136 EKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALE 178
+ +I NL+SEN+ L+ ++K+ E K+E A +E
Sbjct: 442 KDQIANLQSENESLRSQAAAAALEQKVHPE-KIEPDQEVAVVE 567
Score = 52.4 bits (124), Expect = 8e-07
Identities = 57/234 (24%), Positives = 102/234 (43%)
Frame = +1
Query: 217 AALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIE 276
A L+N + +K++ + I+ E+ K + + +++ + E +E
Sbjct: 13 AKLQNALQEMKAQLDEAHAAIIHEREAAKIA---------IEQAPPVIKEVPVVDETKLE 165
Query: 277 NLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQA 336
L ++N++L+ +++E + K E EI N EN+ ++E +LK L+E
Sbjct: 166 LLTNKNEELETEVEELKKKIKEFEESYSEIEN-----ENQARLKEAEEAQLKATQLQETI 330
Query: 337 QRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILEEQAQGKFCDQLKKKCEKVVEG 396
+R LE +S L+SEN+ L Q LEE K E++ E
Sbjct: 331 ER----------------LELSLSNLESENQVLCQKALEE-----------PKNEELFE- 426
Query: 397 RNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALE 450
+KIL+ I NL+SEN+ L+ ++K+ E K+E A +E
Sbjct: 427 ------EIKILKDQIANLQSENESLRSQAAAAALEQKVHPE-KIEPDQEVAVVE 567
Score = 52.0 bits (123), Expect = 1e-06
Identities = 52/211 (24%), Positives = 105/211 (49%), Gaps = 21/211 (9%)
Frame = +1
Query: 311 AALENEVSALKSENKKLKKDILEEQAQRKVAMEG--------------KLEI-SNAFAAL 355
A L+N + +K++ + I+ E+ K+A+E KLE+ +N L
Sbjct: 13 AKLQNALQEMKAQLDEAHAAIIHEREAAKIAIEQAPPVIKEVPVVDETKLELLTNKNEEL 192
Query: 356 ENEVSALKSENKKLKQDI--LEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIEN 413
E EV LK + K+ ++ +E + Q + K+ E+ L++ ++ LE + N
Sbjct: 193 ETEVEELKKKIKEFEESYSEIENENQARL-----KEAEEAQLKATQLQETIERLELSLSN 357
Query: 414 LESENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEVSALKSE--STKLKKDILEEQ 471
LESEN+ L + EE ++ E K+ + + A L++E +L+S+ + L++ + E+
Sbjct: 358 LESENQVLCQKALEEPKNEELFEEIKI-LKDQIANLQSENESLRSQAAAAALEQKVHPEK 534
Query: 472 AQ--IKVAIEEKLEISNAFAALENEVSALKS 500
+ +VA+ EK+++ A +N + +K+
Sbjct: 535 IEPDQEVAVVEKMQVKPRVIA-DNTTAQIKN 624
Score = 50.8 bits (120), Expect = 2e-06
Identities = 56/233 (24%), Positives = 107/233 (45%)
Frame = +1
Query: 82 AALENEVSALISENKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQAVKILEKRIEN 141
A L+N + + ++ + I+ E+ KI + + +++ + E ++E
Sbjct: 13 AKLQNALQEMKAQLDEAHAAIIHEREAAKIA--------IEQAPPVIKEVPVVDETKLEL 168
Query: 142 LESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDILEEQAQ 201
L ++N++L+ +++E ++KI+ + E+S + EN+ ++E +LK L+E +
Sbjct: 169 LTNKNEELETEVEE--LKKKIK---EFEESYSEIENENQARLKEAEEAQLKATQLQETIE 333
Query: 202 RKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGR 261
R LE +S L+SEN+ L Q L+E K E++ E
Sbjct: 334 R----------------LELSLSNLESENQVLCQKALEEP-----------KNEELFE-- 426
Query: 262 NALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALE 314
+KIL+ I NL+SEN+ L+ ++KV E K+E A +E
Sbjct: 427 -----EIKILKDQIANLQSENESLRSQAAAAALEQKVHPE-KIEPDQEVAVVE 567
Score = 48.1 bits (113), Expect = 2e-05
Identities = 47/180 (26%), Positives = 87/180 (48%), Gaps = 4/180 (2%)
Frame = +1
Query: 470 EQAQIKVAIEE-KLEISNAFAALENEVSALKSEIAALQQKCGAGSREGNGDVEVLKAGIS 528
E A+++ A++E K ++ A AA+ +E A K I +E+L
Sbjct: 7 EIAKLQNALQEMKAQLDEAHAAIIHEREAAKIAIEQAPPVIKEVPVVDETKLELLTNKNE 186
Query: 529 DTEKEVNTLKKELVEKEKIVAD--SERKTAVDERKKAAAEARKLLEAAKKIAPEKAVIP- 585
+ E EV LKK++ E E+ ++ +E + + E ++A +A +L E +++ + +
Sbjct: 187 ELETEVEELKKKIKEFEESYSEIENENQARLKEAEEAQLKATQLQETIERLELSLSNLES 366
Query: 586 EPANCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKKENEVSALKSEI 645
E C K E K E L + +KIL+ I NL+SEN+ L+ + +AL+ ++
Sbjct: 367 ENQVLCQKALEEPKNEE--------LFEEIKILKDQIANLQSENESLRSQAAAAALEQKV 522
>TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), partial (7%)
Length = 1040
Score = 52.0 bits (123), Expect = 1e-06
Identities = 70/289 (24%), Positives = 121/289 (41%), Gaps = 7/289 (2%)
Frame = +2
Query: 450 ENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVSALK-SEIAALQQK 508
++E A + + ++++ E + + +E+K+ S A A E E +A E+ A +K
Sbjct: 314 QSEAEAEVEANAESIEEVVAEAEKAQSGVEDKISQSEAEAEAEVEANAKSIEEVVAEAEK 493
Query: 509 CGAGSREGNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEAR 568
+G+ + K S + KE + +L E ++ A+DE K+ EA
Sbjct: 494 AQSGAED--------KISQSVSFKEETNVVGDLPEAQR--------KALDELKRLVQEA- 622
Query: 569 KLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLESE 628
L + AP+ PEP KKK E +K LEK E + E
Sbjct: 623 --LNNHELTAPK----PEPE---------KKKTE------------LKALEKKEEEVSEE 721
Query: 629 NKKLKKENEVSALKSEISALQQKCGAGAREGNGDVEVL--KAGISDTKKEVNRLKKEHVE 686
KKE EV+ K E ++K A E +VEV K + T+++ E +
Sbjct: 722 ----KKEVEVTEEKKEAEVTEEKKEAEVTEEKKEVEVAEEKKEVEGTEEKKEVEVAEEKK 889
Query: 687 EERIVADSERKTAVDERKNAAAEARK----LLEAPKKIAAEVEKQIAKV 731
E ++ + + +E+K K ++E K++ A EK+ A+V
Sbjct: 890 ETEVIKEKKEVEVTEEKKEVEVTEEKKEAEVIEEKKEVDATEEKKEAEV 1036
Score = 36.6 bits (83), Expect = 0.046
Identities = 33/147 (22%), Positives = 67/147 (45%), Gaps = 5/147 (3%)
Frame = +2
Query: 671 SDTKKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAK 730
S+ + EV E +EE A+ + D+ + AEA +EA K +E+ +A+
Sbjct: 317 SEAEAEVEA-NAESIEEVVAEAEKAQSGVEDKISQSEAEAEAEVEANAK---SIEEVVAE 484
Query: 731 VELRQVHLEKQVNERKMKLAFE-----LSKTKEATKRFEAEKKKLLVEKINAESKIKKAQ 785
E Q E ++++ ++F+ + EA ++ E K+L+ E +N
Sbjct: 485 AEKAQSGAEDKISQ---SVSFKEETNVVGDLPEAQRKALDELKRLVQEALNNHELTAPKP 655
Query: 786 ERSESELDKKTADMEKQQAEEQKKLAE 812
E + + + K + ++++ E+KK E
Sbjct: 656 EPEKKKTELKALEKKEEEVSEEKKEVE 736
>TC226006 similar to UP|REMO_SOLTU (P93788) Remorin (pp34), partial (64%)
Length = 991
Score = 48.5 bits (114), Expect = 1e-05
Identities = 56/206 (27%), Positives = 98/206 (47%), Gaps = 24/206 (11%)
Frame = +2
Query: 539 KELVEKEKIVADSERKT-----AVDERKKAAAEARKLLEAAKKIAPEKAVIPEPANCCSK 593
K++ E++ +SE T A + AAEA + EA K + EK+VIP P++ K
Sbjct: 2 KKMTEEQTKTVESESVTPPPAAAAPSQVPPAAEAEHVPEAPKDVTEEKSVIPVPSS-DDK 178
Query: 594 CDELK-----KKCEKVA--------VGRNALRQAVKILEKGIENLES--ENKKLKKENEV 638
DE K +K ++VA + R+A+ V EK + +++ E++K K EN+
Sbjct: 179 PDESKALVLVEKTQEVAEVKPTEGSINRDAVLARV-ATEKRLSLIKAWEESEKSKAENKA 355
Query: 639 SALKSEISALQ--QKCGAGAREGNGDVEVLKAGISDTKKEVNRLKKEH--VEEERIVADS 694
S +SA + +K A + E+ K +K N++ H EE R + ++
Sbjct: 356 HKKLSSVSAWENSKKAAVEADLKKIEEELEKKKAEAAEKIKNKIATIHKEAEERRAIIEA 535
Query: 695 ERKTAVDERKNAAAEARKLLEAPKKI 720
++ + + + AA+ R APKK+
Sbjct: 536 KKGEDLLKAEEQAAKYRATGTAPKKL 613
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 46.6 bits (109), Expect = 4e-05
Identities = 74/309 (23%), Positives = 135/309 (42%), Gaps = 13/309 (4%)
Frame = +3
Query: 143 ESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDILEEQAQR 202
+ E KK K +++ ++K E + K +K++ E E KK KK +
Sbjct: 9 DGEEKKEK----DKKEKKKKENKDKGDKTDVEKVKGKENDGEDDEEKKEKKK------KE 158
Query: 203 KVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDE-QAQGKFCDRLKKKCEKVVEGR 261
K +GK + ++ + + KK K D +D + +GK D G+
Sbjct: 159 KKDKDGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDD-----------GK 305
Query: 262 NALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALK 321
+ + K EKG + + E KK KKD ++E + K + + + + +N+
Sbjct: 306 DEGNKEKKDKEKG--DGDGEEKKEKKDKEKEKKKEKKDKDEETDTLKEKG--KNDEGEDD 473
Query: 322 SENKKLKKDILEEQ----AQRKVAMEGKLEISNAFAALEN-EVSALKSENKKL---KQDI 373
NKK KKD E++ ++K EG+ E S ++ + ++ +K E +K K
Sbjct: 474 EGNKKKKKDKKEKEKDHKKEKKDKEEGEKEDSKVEVSVRDIDIEEIKKEGEKEDKGKDGG 653
Query: 374 LEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKK----LKKDIQEEQ 429
E + + K D+ KK+ +K V G++ + + +K LE N K L+K E+
Sbjct: 654 KEVKEKKKKEDKDKKEKKKKVTGKDKTKDLSTLKQK----LEKINGKIQPLLEKKADIER 821
Query: 430 AQRKIEIEG 438
+++E EG
Sbjct: 822 QIKEVEAEG 848
Score = 39.7 bits (91), Expect = 0.005
Identities = 57/232 (24%), Positives = 90/232 (38%), Gaps = 15/232 (6%)
Frame = +3
Query: 598 KKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAR 657
KKK E G + VK E E+ E + +K KKE + K++ +++K G
Sbjct: 45 KKKKENKDKGDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDKDGKTDAEKIKKKEDDGKD 224
Query: 658 EGNG----------DVEVLKAGISDTKKEVNRLKKEHVE-----EERIVADSERKTAVDE 702
+ D +K D K E N+ KK+ + EE+ + K E
Sbjct: 225 DEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNKEKKDKEKGDGDGEEKKEKKDKEKEKKKE 404
Query: 703 RKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEKQVNERKMKLAFELSKTKEATKR 762
+K+ E L E K E + E ++K K E KE +
Sbjct: 405 KKDKDEETDTLKEKGKNDEGEDD-------------EGNKKKKKDKKEKEKDHKKEKKDK 545
Query: 763 FEAEKKKLLVEKINAESKIKKAQERSESELDKKTADMEKQQAEEQKKLAEDK 814
E EK+ VE + I++ + E E + K D K+ E++KK +DK
Sbjct: 546 EEGEKEDSKVEVSVRDIDIEEI--KKEGEKEDKGKDGGKEVKEKKKKEDKDK 695
Score = 38.5 bits (88), Expect = 0.012
Identities = 76/312 (24%), Positives = 130/312 (41%), Gaps = 8/312 (2%)
Frame = +3
Query: 251 KKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAF 310
KKK E +G + VK E E+ E + +K KK+ K + +GK ++A
Sbjct: 45 KKKKENKDKGDKTDVEKVKGKENDGEDDEEKKEKKKKE--------KKDKDGK---TDAE 191
Query: 311 AALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLK 370
+ E E KK KK ++ KV +GK + K E K K
Sbjct: 192 KIKKKEDDGKDDEEKKEKKKKYDKIDAXKV--KGKEDDG-------------KDEGNKEK 326
Query: 371 QDILEEQAQGKFCDQLK-KKCEKVVEGRNALRQAVKILEKGIENLESE----NKKLKKDI 425
+D + G+ + K K+ EK E ++ + + EKG +N E E NKK KKD
Sbjct: 327 KDKEKGDGDGEEKKEKKDKEKEKKKEKKDKDEETDTLKEKG-KNDEGEDDEGNKKKKKDK 503
Query: 426 QEEQAQRKIEIEGKLEISNAFAALENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEIS 485
+E++ K E + K E + +E V + E K + + ++ ++EK +
Sbjct: 504 KEKEKDHKKEKKDKEEGEKEDSKVEVSVRDIDIEEIKKEGEKEDKGKDGGKEVKEKKKKE 683
Query: 486 NAFAALENEVSALK---SEIAALQQKCGAGSREGNGDVEVLKAGISDTEKEVNTLKKELV 542
+ + + K +++ L+QK + NG ++ L +D E+++ V
Sbjct: 684 DKDKKEKKKKVTGKDKTKDLSTLKQKL----EKINGKIQPLLEKKADIERQIKE-----V 836
Query: 543 EKEKIVADSERK 554
E E V + E K
Sbjct: 837 EAEGHVVNEENK 872
>TC216950
Length = 873
Score = 45.8 bits (107), Expect = 8e-05
Identities = 43/151 (28%), Positives = 69/151 (45%)
Frame = +2
Query: 666 LKAGISDTKKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVE 725
LK +T K V ++ VEE + E + ++ R E RK L E E
Sbjct: 221 LKLIEEETAKRVEEAIRKRVEES--LKSEEVQVEIERRLE---EGRKRLNDEVAAQLEKE 385
Query: 726 KQIAKVELRQVHLEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQ 785
K+ A +E +Q K+ RK K E + K EA++++ L ++ E + K
Sbjct: 386 KEAALIEAKQ----KEEQARKEKEDLERMLEENRRKVEEAQRREALEQQRREEERYK--- 544
Query: 786 ERSESELDKKTADMEKQQAEEQKKLAEDKLL 816
E E + K+ A +K+ EEQ++L + KLL
Sbjct: 545 ELEEMQRQKEEAMRKKKHEEEQERLNQIKLL 637
Score = 37.0 bits (84), Expect = 0.035
Identities = 35/134 (26%), Positives = 67/134 (49%), Gaps = 12/134 (8%)
Frame = +2
Query: 114 QLKKCEKVVEGRNALRQAVKILEKRIENLESENKKLKKDIQEEQAQRKI--EIEGKLEKS 171
QL+ K++E A R I ++ E+L+SE +++ + + E+ ++++ E+ +LEK
Sbjct: 206 QLEAELKLIEEETAKRVEEAIRKRVEESLKSEEVQVEIERRLEEGRKRLNDEVAAQLEKE 385
Query: 172 NAFAALENEVSALKSENKKLKKDILE----------EQAQRKVAMEGKLEISNAFAALEN 221
A +E + K E + +K+ LE E+AQR+ A+E + + LE
Sbjct: 386 KEAALIE---AKQKEEQARKEKEDLERMLEENRRKVEEAQRREALEQQRREEERYKELE- 553
Query: 222 EVSALKSENKKLKQ 235
E+ K E + K+
Sbjct: 554 EMQRQKEEAMRKKK 595
>BU550864
Length = 636
Score = 45.4 bits (106), Expect = 1e-04
Identities = 45/180 (25%), Positives = 78/180 (43%), Gaps = 15/180 (8%)
Frame = -3
Query: 125 RNALRQAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSAL 184
R L + + EKR + L++E +K +E ++ IEG +A LE + L
Sbjct: 634 RAELETSRLLAEKRKQELDTE----RKCAEELNETMQMAIEGHARILEQYADLEEKHIQL 467
Query: 185 KSENKKLKKDI--LEEQAQRKVAMEGK---------LEISNAFAALENEVSALKSENKKL 233
++K++ I ++E A R + G EIS A E E+ L+ EN+ L
Sbjct: 466 LDRHRKIQDGIDDVKEAASRAAGVRGADSKFINALAAEISALKAEKEKEMEILRDENRGL 287
Query: 234 KQDILDE----QAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDI 289
+ + D QA G+ +L + E V+ + A + K + ++ KK K+I
Sbjct: 286 RAQLKDTAEAVQAAGELLVQLNEAEEAVITAQKRAMDAEQEAAKAYKQIDKLKKKHDKEI 107
Score = 41.6 bits (96), Expect = 0.001
Identities = 51/194 (26%), Positives = 88/194 (45%), Gaps = 13/194 (6%)
Frame = -3
Query: 220 ENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCEKVVEGR-NALRQAVKILEKGIENL 278
E E S L +E +K + D + K + L + + +EG L Q + EK I+ L
Sbjct: 628 ELETSRLLAEKRKQELD-----TERKCAEELNETMQMAIEGHARILEQYADLEEKHIQLL 464
Query: 279 ESENKKLKKDIQE--EHAQRKVEIEGK---------LEISNAFAALENEVSALKSENKKL 327
+ ++K++ I + E A R + G EIS A E E+ L+ EN+ L
Sbjct: 463 D-RHRKIQDGIDDVKEAASRAAGVRGADSKFINALAAEISALKAEKEKEMEILRDENRGL 287
Query: 328 KKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILEEQAQG-KFCDQL 386
+ + + + A E ++++ A A+ ++A +K D +E A+ K D+L
Sbjct: 286 RAQLKDTAEAVQAAGELLVQLNEAEEAV---ITA-----QKRAMDAEQEAAKAYKQIDKL 131
Query: 387 KKKCEKVVEGRNAL 400
KKK +K + N L
Sbjct: 130 KKKHDKEISTLNKL 89
>TC206766 similar to PIR|T47576|T47576 FKBP12 interacting protein (FIP37) -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(74%)
Length = 1952
Score = 45.1 bits (105), Expect = 1e-04
Identities = 60/296 (20%), Positives = 125/296 (41%), Gaps = 16/296 (5%)
Frame = +3
Query: 444 NAFAALENEVSALKSESTKLKKDILEEQAQIKVAIEEKLEISNAFAALENEVSALKSEIA 503
+ A +NE+ A KSE + + + + E + N AL++ +L+ ++
Sbjct: 402 DTLATCQNELEAAKSEIQSWHSTLKNQPSILAGITPEPKMLINYLQALKSSEESLREQLE 581
Query: 504 ALQQKCGA-----GSREGNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVAD-------S 551
++K A RE ++ LK+ + D + + LK ++ +++ D +
Sbjct: 582 KAKKKEAAFIVTFAKRE--QEIAELKSAVRDLKVQ---LKPPSMQSRRLLLDPAVHEEFT 746
Query: 552 ERKTAVDERKKAAAEARKLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKVAVGRNAL 611
K V+E+ K E + + AA P+ + +KC L+++ E+ +G A
Sbjct: 747 RLKNLVEEKDKKVKELQDNI-AAVSFTPQSKM---GKMLMAKCRTLQEENEE--IGNQAS 908
Query: 612 RQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAREGNGDVEVLKAGIS 671
+ L + +S+N +L+ + E L+ + L N V +L+ +
Sbjct: 909 EGKMHELGMKLALQKSQNSQLRSQFE--GLQKHMEGLTN----DVERSNEMVLMLQEKLE 1070
Query: 672 DTKKEVNRLKKE----HVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAE 723
D +E+ RLK E ++E++R+ ++K D+R + + L + A E
Sbjct: 1071DRDREIQRLKHELQQKNLEDQRLKHKLQQKNLEDQRLKHELQQKNLEDGNDNNAGE 1238
>TC228989 similar to UP|Q9LSS5 (Q9LSS5) Myosin heavy chain-like, partial
(23%)
Length = 884
Score = 44.7 bits (104), Expect = 2e-04
Identities = 52/201 (25%), Positives = 91/201 (44%), Gaps = 12/201 (5%)
Frame = +1
Query: 449 LENEVSALKSESTKLKKDILEEQAQIKVAIEE---KLEISNAFAALENEVSALKSEIAAL 505
LE EV +LKSE +L+ I E A+ K E+ ++I NA+ +E +KSE
Sbjct: 37 LEAEVVSLKSEVEQLRSAI--ETAETKYQEEQIRSMVQIRNAYELVEQ----IKSESGKR 198
Query: 506 QQKCGAGSREGNGDVEVLKAGISDTEKEVNTLKKE-----LVEKEKIVADSERKTAVDER 560
+ + A ++ D+E LKA + D E E+ + +E L +E + + +E K + +
Sbjct: 199 ECELEAELKKKKADIEELKANLMDKETELQGIVEENDNLNLKLEESMSSKNEHKLKREIK 378
Query: 561 KKAAAEARKLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKVAV----GRNALRQAVK 616
+ A A + K +++ E S+ + K E+VA + A +A+
Sbjct: 379 RLAECVAELKADMMDKETTLQSISEENEMLKSEINNSGKAREEVAAEVEGAKAAEHEALM 558
Query: 617 ILEKGIENLESENKKLKKENE 637
L +E + NKK + E
Sbjct: 559 KLGIVMEEADKSNKKAARVAE 621
Score = 36.6 bits (83), Expect = 0.046
Identities = 48/224 (21%), Positives = 95/224 (41%), Gaps = 21/224 (9%)
Frame = +1
Query: 610 ALRQAVKILEKGIENLESENKK--------LKKENE-VSALKSEISALQQKCGAGAREGN 660
+L+ V+ L IE E++ ++ ++ E V +KSE + + A ++
Sbjct: 55 SLKSEVEQLRSAIETAETKYQEEQIRSMVQIRNAYELVEQIKSESGKRECELEAELKKKK 234
Query: 661 GDVEVLKAGISDTKKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKI 720
D+E LKA + D + E+ + +E+ + + E + E ++L E ++
Sbjct: 235 ADIEELKANLMDKETELQGIVEEN---DNLNLKLEESMSSKNEHKLKREIKRLAECVAEL 405
Query: 721 AAEV---EKQIAKVELRQVHLEKQVNERKMKLAFELSKTKEATKRFEAEK-KKLLVEKIN 776
A++ E + + L+ ++N K E++ E K E E KL +
Sbjct: 406 KADMMDKETTLQSISEENEMLKSEIN-NSGKAREEVAAEVEGAKAAEHEALMKLGIVMEE 582
Query: 777 AESKIKKAQERSESELDKKTADME--------KQQAEEQKKLAE 812
A+ KKA +E + A++E K Q+++ +K AE
Sbjct: 583 ADKSNKKAARVAEQLEAAQAANLEMEAELRRLKVQSDQWRKAAE 714
>BG239031
Length = 367
Score = 43.9 bits (102), Expect = 3e-04
Identities = 37/114 (32%), Positives = 62/114 (53%), Gaps = 12/114 (10%)
Frame = +1
Query: 400 LRQAVKILEKGIENLESENKKLKKDIQEEQAQRKIEIEGKLEISNAFAALENEVSALKSE 459
L + +KIL+ I NLESEN+ L++ ++K+ E K+E ++ A+LE +V K E
Sbjct: 25 LFEEIKILKDQIANLESENESLRRQAAAVAFEQKVHPE-KIESDHSAASLEQKVQPEKIE 201
Query: 460 ----STKLKKDILEEQAQ---IKVAIE-----EKLEISNAFAALENEVSALKSE 501
+T L++ + E+ + A+E EK+E ++ AALE +V K E
Sbjct: 202 PGHSATALEQKVNPEKIEPVHSATALEQKVHPEKIEPDHSAAALEQKVHPEKIE 363
Score = 42.4 bits (98), Expect = 8e-04
Identities = 36/115 (31%), Positives = 61/115 (52%), Gaps = 13/115 (11%)
Frame = +1
Query: 128 LRQAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSE 187
L + +KIL+ +I NLESEN+ L++ ++K+ E K+E ++ A+LE +V K E
Sbjct: 25 LFEEIKILKDQIANLESENESLRRQAAAVAFEQKVHPE-KIESDHSAASLEQKVQPEKIE 201
Query: 188 --------NKKLKKDILE-----EQAQRKVAMEGKLEISNAFAALENEVSALKSE 229
+K+ + +E ++KV E K+E ++ AALE +V K E
Sbjct: 202 PGHSATALEQKVNPEKIEPVHSATALEQKVHPE-KIEPDHSAAALEQKVHPEKIE 363
Score = 40.4 bits (93), Expect = 0.003
Identities = 37/115 (32%), Positives = 60/115 (52%), Gaps = 13/115 (11%)
Frame = +1
Query: 264 LRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALKSE 323
L + +KIL+ I NLESEN+ L++ ++KV E K+E ++ A+LE +V K E
Sbjct: 25 LFEEIKILKDQIANLESENESLRRQAAAVAFEQKVHPE-KIESDHSAASLEQKVQPEKIE 201
Query: 324 --------NKKLKKDILE-----EQAQRKVAMEGKLEISNAFAALENEVSALKSE 365
+K+ + +E ++KV E K+E ++ AALE +V K E
Sbjct: 202 PGHSATALEQKVNPEKIEPVHSATALEQKVHPE-KIEPDHSAAALEQKVHPEKIE 363
Score = 37.4 bits (85), Expect = 0.027
Identities = 40/153 (26%), Positives = 66/153 (42%)
Frame = +1
Query: 35 LRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEGKLEISNTFAALENEVSALISE 94
L + +KIL+ I NLESEN+ L++ ++KV E K+E ++ A+LE +V
Sbjct: 25 LFEEIKILKDQIANLESENESLRRQAAAVAFEQKVHPE-KIESDHSAASLEQKVQP---- 189
Query: 95 NKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQAVKILEKRIENLESENKKLKKDIQ 154
EK+ G +A K+ ++IE + S L++ +
Sbjct: 190 ------------------------EKIEPGHSATALEQKVNPEKIEPVHSAT-ALEQKVH 294
Query: 155 EEQAQRKIEIEGKLEKSNAFAALENEVSALKSE 187
E K+E ++ AALE +V K E
Sbjct: 295 PE----------KIEPDHSAAALEQKVHPEKIE 363
Score = 35.8 bits (81), Expect = 0.079
Identities = 31/114 (27%), Positives = 56/114 (48%), Gaps = 4/114 (3%)
Frame = +1
Query: 611 LRQAVKILEKGIENLESENKKLKKENEVSALKSEI--SALQQKCGAGAREGNGDVEVLKA 668
L + +KIL+ I NLESEN+ L+++ A + ++ ++ A + E E ++
Sbjct: 25 LFEEIKILKDQIANLESENESLRRQAAAVAFEQKVHPEKIESDHSAASLEQKVQPEKIEP 204
Query: 669 GISDT--KKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKI 720
G S T +++VN K E V + ++ +AAA +K+ P+KI
Sbjct: 205 GHSATALEQKVNPEKIEPVHSATALEQKVHPEKIEPDHSAAALEQKV--HPEKI 360
>TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-infected
erythrocyte surface antigen, partial (5%)
Length = 858
Score = 42.0 bits (97), Expect = 0.001
Identities = 66/248 (26%), Positives = 107/248 (42%), Gaps = 11/248 (4%)
Frame = +1
Query: 440 LEISNAFAALENEVSALKS--ESTKLKKDILEEQAQIKVAIEEKLEISNAFAALEN-EVS 496
+E A + +E+++S S E T + D+ E Q K A++E ++ AL N E++
Sbjct: 190 IEAEKAQSGVEDKISQSVSFKEETNVVGDLPEAQ---KKALDELKKLVQE--ALNNHELT 354
Query: 497 ALKSEIA----ALQQKCGAGSREGNGDVEVLKAGISDTEKEVNTLKKELV----EKEKIV 548
A K E A ++K EG + EV++ E EV KKE+ +KE V
Sbjct: 355 APKPEPEKKKPAAEKKEEVEVTEGKKEAEVIE---EKKEVEVTEEKKEIEVTEEKKEAEV 525
Query: 549 ADSERKTAVDERKKAAAEARKLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCEKVAVGR 608
+ +++ V E KK + EA K ++ + E +E K
Sbjct: 526 IEEKKEVEVTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKK---------- 675
Query: 609 NALRQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAREGNGDVEVLKA 668
+A I+E E E E + KKE EV+ K E+ +++K E +VEV
Sbjct: 676 ----EAEVIVE---EKKEVEVTEEKKEVEVTEGKKEVEVIEEKKETEVTEEKKEVEV--- 825
Query: 669 GISDTKKE 676
+ + KKE
Sbjct: 826 EVREEKKE 849
>TC214570 homologue to PIR|T05766|T05766 peptidylprolyl isomerase M4E13.20 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(13%)
Length = 944
Score = 41.2 bits (95), Expect = 0.002
Identities = 45/153 (29%), Positives = 63/153 (40%), Gaps = 1/153 (0%)
Frame = +2
Query: 96 KKLKQDILEE-QAQGKICDQLKKCEKVVEGRNALRQAVKILEKRIENLESENKKLKKDIQ 154
KK+K D +EE + Q K+ K +KV E + +E +E E + KK K
Sbjct: 389 KKVKVDEVEEVEKQEKV-----KVKKVDE---------ETVEVEVEKKEKKKKKKKNKEN 526
Query: 155 EEQAQRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEISN 214
E A E K +K + ++ KSE KK KK E A GK + SN
Sbjct: 527 SEAASSDEEKTEKKKKKHKDKVEDSSPELDKSEKKKKKKKDKEAAAATAEISNGKEDDSN 706
Query: 215 AFAALENEVSALKSENKKLKQDILDEQAQGKFC 247
A + S K KK +D +E + FC
Sbjct: 707 A------DKSEKKKHKKKKNKDADEE*RKAYFC 787
Score = 37.7 bits (86), Expect = 0.021
Identities = 45/154 (29%), Positives = 69/154 (44%), Gaps = 1/154 (0%)
Frame = +2
Query: 231 KKLKQDILDE-QAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDI 289
KK+K D ++E + Q K ++KK E+ VE ++ K +K EN E+ +
Sbjct: 389 KKVKVDEVEEVEKQEKV--KVKKVDEETVEVEVEKKEKKKKKKKNKENSEAASSD----- 547
Query: 290 QEEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQRKVAMEGKLEIS 349
+E+ ++K + + K+E S+ KSE KK KK E A GK + S
Sbjct: 548 EEKTEKKKKKHKDKVEDSSP--------ELDKSEKKKKKKKDKEAAAATAEISNGKEDDS 703
Query: 350 NAFAALENEVSALKSENKKLKQDILEEQAQGKFC 383
NA + S K KK +D EE + FC
Sbjct: 704 NA------DKSEKKKHKKKKNKDADEE*RKAYFC 787
Score = 29.3 bits (64), Expect = 7.4
Identities = 23/87 (26%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Frame = +2
Query: 730 KVELRQVH-LEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQERS 788
KV++ +V +EKQ + K+ E + + K + +KKK A S +K +++
Sbjct: 392 KVKVDEVEEVEKQEKVKVKKVDEETVEVEVEKKEKKKKKKKNKENSEAASSDEEKTEKKK 571
Query: 789 ESELDK-KTADMEKQQAEEQKKLAEDK 814
+ DK + + E ++E++KK +DK
Sbjct: 572 KKHKDKVEDSSPELDKSEKKKKKKKDK 652
>CO983030
Length = 505
Score = 40.0 bits (92), Expect = 0.004
Identities = 29/129 (22%), Positives = 60/129 (46%)
Frame = +3
Query: 681 KKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEK 740
KK+ ++++ ++K ++K + +K + KK + +K+ K + ++ +K
Sbjct: 9 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 188
Query: 741 QVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQERSESELDKKTADME 800
+ ++K K + K K+ K+ + +KKK K KIKK Q++ + KK
Sbjct: 189 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKNKKKKXXKIKKTQKKKXXKT*KKKXKTL 368
Query: 801 KQQAEEQKK 809
K+ KK
Sbjct: 369 KKNFTNLKK 395
Score = 36.6 bits (83), Expect = 0.046
Identities = 24/108 (22%), Positives = 55/108 (50%)
Frame = +1
Query: 702 ERKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEKQVNERKMKLAFELSKTKEATK 761
++K + +K + KK + +K+ K + ++ +K+ ++K K + K K+ K
Sbjct: 4 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 183
Query: 762 RFEAEKKKLLVEKINAESKIKKAQERSESELDKKTADMEKQQAEEQKK 809
+ + +KKK +K + K KK +++ + + KKT + + ++ KK
Sbjct: 184 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKTKKKKXXK*KKHKK 327
Score = 33.9 bits (76), Expect = 0.30
Identities = 31/152 (20%), Positives = 69/152 (45%), Gaps = 1/152 (0%)
Frame = +3
Query: 136 EKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSENKKLKKDI 195
+K+ + + + KK KK ++++ ++K + + K +K + + K + KK KK
Sbjct: 3 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 182
Query: 196 LEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKFCDRLKKKCE 255
+++ ++K + K + + + K +NKK K + + + K KKK +
Sbjct: 183 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKNKKKKXXKIKKTQKKKXXKT*KKKXK 362
Query: 256 KVVEGRNALRQAVKILE-KGIENLESENKKLK 286
+ + L++ ++ GI NL + K K
Sbjct: 363 TLKKNFTNLKKXXTLINXXGIXNLMLQ*NKQK 458
Score = 32.7 bits (73), Expect = 0.67
Identities = 26/121 (21%), Positives = 56/121 (45%)
Frame = +2
Query: 531 EKEVNTLKKELVEKEKIVADSERKTAVDERKKAAAEARKLLEAAKKIAPEKAVIPEPANC 590
+K+ KK+ +K+K ++K ++KK + +K + KK +K +
Sbjct: 14 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 193
Query: 591 CSKCDELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQ 650
K + KKK +K + ++ K +K + + +NKK K+ + +K +I +++
Sbjct: 194 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKQKKKKXKNKKNTKKKKXXNMKKKIXNIKK 373
Query: 651 K 651
K
Sbjct: 374 K 376
Score = 32.0 bits (71), Expect = 1.1
Identities = 32/157 (20%), Positives = 72/157 (45%), Gaps = 9/157 (5%)
Frame = +2
Query: 674 KKEVNRLKKEHVEEERIVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVEL 733
KK+ + KK+ ++++ ++K ++K + +K + KK + +K+ K +
Sbjct: 44 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 223
Query: 734 RQVHLEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQER-----S 788
++ +K+ ++K K + + K+ K + KKK + N + KI +++
Sbjct: 224 KKKKKKKKKKKKKKKKKKKKKQKKKKXKNKKNTKKK---KXXNMKKKIXNIKKKFYKP*K 394
Query: 789 ESELDKKTADMEKQQAEEQKKLAE----DKLLLLGDS 821
+DK+ + + +Q K AE KL+ +G S
Sbjct: 395 IXNIDKQXWNXKSHATIKQTKNAELLALKKLVWIGAS 505
Score = 29.6 bits (65), Expect = 5.7
Identities = 29/145 (20%), Positives = 64/145 (44%), Gaps = 1/145 (0%)
Frame = +3
Query: 279 ESENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQR 338
+ + KK KK +++ ++K + + K + + + K + KK KK +++ ++
Sbjct: 24 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 203
Query: 339 KVAMEGKLEISNAFAALENEVSALKSENKKLKQDILEEQAQGKFCDQLKKKCEKVVEGRN 398
K + K + + + K +NKK K +++ + K KKK + + +
Sbjct: 204 KKKKKKKKKKKKKKKKKKKKKKKKKKKNKKKKXXKIKKTQKKKXXKT*KKKXKTLKKNFT 383
Query: 399 ALRQAVKILE-KGIENLESENKKLK 422
L++ ++ GI NL + K K
Sbjct: 384 NLKKXXTLINXXGIXNLMLQ*NKQK 458
>CF921229
Length = 680
Score = 38.5 bits (88), Expect = 0.012
Identities = 38/145 (26%), Positives = 75/145 (51%), Gaps = 7/145 (4%)
Frame = -3
Query: 683 EHVEEERIVADSERKTAVD---ERKNAAAEARK----LLEAPKKIAAEVEKQIAKVELRQ 735
E +EE++ V +E K ++ E+K A + K + E K++ EK+ A+V + +
Sbjct: 666 EVIEEKKEVEVTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKKEAEVIVEE 487
Query: 736 VHLEKQVNERKMKLAFELSKTKEATKRFEAEKKKLLVEKINAESKIKKAQERSESELDKK 795
+K+V + K E+++ K+ + E EKK+ V + E +++ +E+ ESE+ ++
Sbjct: 486 ---KKEVEVTEEKKEVEVTEGKKEVEVIE-EKKETEVTEEKKEVEVEVREEKKESEVKEE 319
Query: 796 TADMEKQQAEEQKKLAEDKLLLLGD 820
EK Q +++ + LLGD
Sbjct: 318 ----EKGQEVVPEEVEIWGIPLLGD 256
Score = 32.7 bits (73), Expect = 0.67
Identities = 38/153 (24%), Positives = 65/153 (41%)
Frame = -3
Query: 543 EKEKIVADSERKTAVDERKKAAAEARKLLEAAKKIAPEKAVIPEPANCCSKCDELKKKCE 602
+KE V + +++ V E KK + EA K ++ + E +E K
Sbjct: 678 KKEAEVIEEKKEVEVTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKK---- 511
Query: 603 KVAVGRNALRQAVKILEKGIENLESENKKLKKENEVSALKSEISALQQKCGAGAREGNGD 662
+A I+E E E E + KKE EV+ K E+ +++K E +
Sbjct: 510 ----------EAEVIVE---EKKEVEVTEEKKEVEVTEGKKEVEVIEEKKETEVTEEKKE 370
Query: 663 VEVLKAGISDTKKEVNRLKKEHVEEERIVADSE 695
VEV + + KKE + +K+E +E + + E
Sbjct: 369 VEV---EVREEKKE-SEVKEEEKGQEVVPEEVE 283
>TC216502 similar to GB|AAF40452.1|7211981|F13M7 ESTs gb|N65605, gb|N38087,
gb|T20485, gb|T13726, gb|N38339, gb|F15440 and gb|N97201
come from this gene. {Arabidopsis thaliana;} , partial
(94%)
Length = 1551
Score = 38.1 bits (87), Expect = 0.016
Identities = 34/98 (34%), Positives = 54/98 (54%), Gaps = 4/98 (4%)
Frame = +2
Query: 80 TFAALENEVSALISENKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQAVKILE-KR 138
T +E L E K KQ L E+A+ K ++ K+ E+ E + +R ++LE KR
Sbjct: 608 TNTKIEAPKPLLTPEEMKAKQQELRERARKKKEEEEKRMERERE-KERIRIGKELLEAKR 784
Query: 139 IENLESENKKL---KKDIQEEQAQRKIEIEGKLEKSNA 173
IE E+E K+L KK +EE+ + + +I+ KLE+ A
Sbjct: 785 IEE-ENERKRLLALKKVEKEEEKRAREKIKKKLEEDKA 895
Score = 36.2 bits (82), Expect = 0.060
Identities = 27/103 (26%), Positives = 51/103 (49%), Gaps = 6/103 (5%)
Frame = +2
Query: 714 LEAPKKIAAEVEKQIAKVELRQ------VHLEKQVNERKMKLAFELSKTKEATKRFEAEK 767
+EAPK + E + + ELR+ EK++ + K + K KR E E
Sbjct: 620 IEAPKPLLTPEEMKAKQQELRERARKKKEEEEKRMEREREKERIRIGKELLEAKRIEEEN 799
Query: 768 KKLLVEKINAESKIKKAQERSESELDKKTADMEKQQAEEQKKL 810
++ +++ A K++K +E+ E KK +E+ +AE +++L
Sbjct: 800 ER---KRLLALKKVEKEEEKRAREKIKK--KLEEDKAERRRRL 913
Score = 30.8 bits (68), Expect = 2.5
Identities = 24/94 (25%), Positives = 45/94 (47%), Gaps = 2/94 (2%)
Frame = +2
Query: 690 IVADSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKV--ELRQVHLEKQVNERKM 747
++ E K E + A + ++ E K++ E EK+ ++ EL + ++ NERK
Sbjct: 638 LLTPEEMKAKQQELRERARKKKE--EEEKRMEREREKERIRIGKELLEAKRIEEENERKR 811
Query: 748 KLAFELSKTKEATKRFEAEKKKLLVEKINAESKI 781
LA + + +E + E KKKL +K ++
Sbjct: 812 LLALKKVEKEEEKRAREKIKKKLEEDKAERRRRL 913
Score = 30.8 bits (68), Expect = 2.5
Identities = 23/76 (30%), Positives = 39/76 (51%)
Frame = +2
Query: 501 EIAALQQKCGAGSREGNGDVEVLKAGISDTEKEVNTLKKELVEKEKIVADSERKTAVDER 560
E+ A QQ+ +R+ + E K + EKE + KEL+E ++I ++ERK + +
Sbjct: 653 EMKAKQQELRERARKKKEEEE--KRMEREREKERIRIGKELLEAKRIEEENERKRLLALK 826
Query: 561 KKAAAEARKLLEAAKK 576
K E ++ E KK
Sbjct: 827 KVEKEEEKRAREKIKK 874
>TC230269 weakly similar to UP|WSC2_YEAST (P53832) Cell wall integrity and
stress response component 2 precursor, partial (6%)
Length = 696
Score = 37.7 bits (86), Expect = 0.021
Identities = 38/136 (27%), Positives = 61/136 (43%), Gaps = 2/136 (1%)
Frame = +2
Query: 108 QGKICDQLKKCEKVVEGRNALRQAVKILEKRIE-NLESENKKLKKDIQEEQAQRKIEIEG 166
+ K+ D++++ EK+ + + K+ E+ +E +E + KK KK +E ++ E
Sbjct: 35 KAKVVDEVEELEKLEKVK-----VKKVDEETVEVEVEKKEKKKKKKKDKENSEVASSDEE 199
Query: 167 KLEKSNAFAALENEVSAL-KSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSA 225
K EK +E+ L KSE KK KK + A EISN N +
Sbjct: 200 KTEKKKNKNKIEDGSPELDKSEKKKKKK--------KDKAAAAAAEISNGKVDDSNADKS 355
Query: 226 LKSENKKLKQDILDEQ 241
K +NKK K E+
Sbjct: 356 EKKKNKKKKNKDAKEE 403
Score = 33.5 bits (75), Expect = 0.39
Identities = 40/139 (28%), Positives = 58/139 (40%), Gaps = 8/139 (5%)
Frame = +2
Query: 253 KCEKVVEGRNALRQAVKILEKGIEN------LESENKKLKKDIQEEHAQRKVEIEGKLEI 306
K KVV+ L + K+ K ++ +E + KK KK +E+++ E K E
Sbjct: 32 KKAKVVDEVEELEKLEKVKVKKVDEETVEVEVEKKEKKKKKKKDKENSEVASSDEEKTEK 211
Query: 307 SNAFAALENEVSAL-KSENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSE 365
+E+ L KSE KK KK + A EISN N + K +
Sbjct: 212 KKNKNKIEDGSPELDKSEKKKKKK--------KDKAAAAAAEISNGKVDDSNADKSEKKK 367
Query: 366 NKKLK-QDILEEQAQGKFC 383
NKK K +D EE + C
Sbjct: 368 NKKKKNKDAKEE*RKAYVC 424
>TC229023
Length = 1337
Score = 37.4 bits (85), Expect = 0.027
Identities = 31/127 (24%), Positives = 58/127 (45%), Gaps = 4/127 (3%)
Frame = +1
Query: 251 KKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQEEHAQRKVEIEGKLEISNAF 310
K E E +N L A+ +E IE L+S+ K + + + Q V E LE++
Sbjct: 517 KASSEASQEQQNMLYTAIWDMEILIEELKSKVAKAETNKESAAEQYFVLSETNLELNKEL 696
Query: 311 AALENEVSALKSE----NKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSEN 366
L + +LK+ + E + K+ M+ +++++ + N++ ALK EN
Sbjct: 697 DLLRSRTVSLKTSLDQASNTRSSRAKEIDCKAKLIMDMVMQLASERERINNQLHALKQEN 876
Query: 367 KKLKQDI 373
K L + +
Sbjct: 877 KHLVEKL 897
Score = 35.4 bits (80), Expect = 0.10
Identities = 28/119 (23%), Positives = 55/119 (45%), Gaps = 4/119 (3%)
Frame = +1
Query: 123 EGRNALRQAVKILEKRIENLESENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVS 182
E +N L A+ +E IE L+S+ K + + + Q + E LE + L +
Sbjct: 541 EQQNMLYTAIWDMEILIEELKSKVAKAETNKESAAEQYFVLSETNLELNKELDLLRSRTV 720
Query: 183 ALKSE----NKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDI 237
+LK+ + E + K+ M+ +++++ + N++ ALK ENK L + +
Sbjct: 721 SLKTSLDQASNTRSSRAKEIDCKAKLIMDMVMQLASERERINNQLHALKQENKHLVEKL 897
>TC229451 similar to UP|Q9FH68 (Q9FH68) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K16E1, partial (80%)
Length = 850
Score = 37.4 bits (85), Expect = 0.027
Identities = 41/162 (25%), Positives = 72/162 (44%), Gaps = 3/162 (1%)
Frame = +2
Query: 73 GKLEISNTFAALENEVSALISENKKLKQDILEEQAQGKICDQLKKCEKVVEGRNALRQAV 132
G + +S A L +++ + K++Q LE DQ+ + ++E +L V
Sbjct: 236 GPVVVSTVGATLVVVLASSLYSMAKIQQRTLEAGIVNPT-DQVLMSKHMLEA--SLMGFV 406
Query: 133 KILEKRIENLES---ENKKLKKDIQEEQAQRKIEIEGKLEKSNAFAALENEVSALKSENK 189
L I+ L E + L+K ++ + Q + +GK S AL E++ LKS+ K
Sbjct: 407 LFLSLMIDRLHHYIRELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIK 586
Query: 190 KLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENK 231
KL+ + + +Q K LE EV ALK +++
Sbjct: 587 KLESECEAKGSQAK--------------TLETEVEALKKQSE 670
Score = 36.2 bits (82), Expect = 0.060
Identities = 31/126 (24%), Positives = 56/126 (43%), Gaps = 7/126 (5%)
Frame = +2
Query: 368 KLKQDILEEQAQGKFCDQLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQE 427
+L I E ++ K + +KK+ +G+N + K L + I L+S+ KKL+ + +
Sbjct: 431 RLHHYIRELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEA 610
Query: 428 EQAQRKIEIEGKLEISNAFAALENEVSALKSES-------TKLKKDILEEQAQIKVAIEE 480
+ +Q K LE EV ALK +S +L +D ++Q++ +
Sbjct: 611 KGSQAK--------------TLETEVEALKKQSEGFLMEYDRLLEDNQSLRSQLQAIDQS 748
Query: 481 KLEISN 486
L N
Sbjct: 749 PLNFDN 766
Score = 33.9 bits (76), Expect = 0.30
Identities = 29/109 (26%), Positives = 54/109 (48%), Gaps = 11/109 (10%)
Frame = +2
Query: 323 ENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILEEQAQGKF 382
E + L+K + + Q + +GK S AL E++ LKS+ KKL+ + + +Q K
Sbjct: 452 ELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEAKGSQAKT 631
Query: 383 ----CDQLKKKCE-------KVVEGRNALRQAVKILEKGIENLESENKK 420
+ LKK+ E +++E +LR ++ +++ L +NKK
Sbjct: 632 LETEVEALKKQSEGFLMEYDRLLEDNQSLRSQLQAIDQ--SPLNFDNKK 772
Score = 33.9 bits (76), Expect = 0.30
Identities = 24/94 (25%), Positives = 45/94 (47%)
Frame = +2
Query: 232 KLKQDILDEQAQGKFCDRLKKKCEKVVEGRNALRQAVKILEKGIENLESENKKLKKDIQE 291
+L I + ++ K + +KK+ +G+N + K L + I L+S+ KKL+ + +
Sbjct: 431 RLHHYIRELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEA 610
Query: 292 EHAQRKVEIEGKLEISNAFAALENEVSALKSENK 325
+ +Q K LE EV ALK +++
Sbjct: 611 KGSQAK--------------TLETEVEALKKQSE 670
Score = 33.5 bits (75), Expect = 0.39
Identities = 31/101 (30%), Positives = 46/101 (44%), Gaps = 2/101 (1%)
Frame = +2
Query: 281 ENKKLKKDIQEEHAQRKVEIEGKLEISNAFAALENEVSALKSENKKLKKDILEEQAQRKV 340
E + L+K ++ Q + +GK S AL E++ LKS+ KKL+ + + +Q K
Sbjct: 452 ELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEAKGSQAK- 628
Query: 341 AMEGKLEISNAFAALENEVSALK--SENKKLKQDILEEQAQ 379
LE EV ALK SE ++ D L E Q
Sbjct: 629 -------------TLETEVEALKKQSEGFLMEYDRLLEDNQ 712
Score = 33.5 bits (75), Expect = 0.39
Identities = 29/109 (26%), Positives = 54/109 (48%), Gaps = 11/109 (10%)
Frame = +2
Query: 187 ENKKLKKDILEEQAQRKVAMEGKLEISNAFAALENEVSALKSENKKLKQDILDEQAQGKF 246
E + L+K + + Q + +GK S AL E++ LKS+ KKL+ + + +Q K
Sbjct: 452 ELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEAKGSQAKT 631
Query: 247 ----CDRLKKKCE-------KVVEGRNALRQAVKILEKGIENLESENKK 284
+ LKK+ E +++E +LR ++ +++ L +NKK
Sbjct: 632 LETEVEALKKQSEGFLMEYDRLLEDNQSLRSQLQAIDQ--SPLNFDNKK 772
Score = 32.7 bits (73), Expect = 0.67
Identities = 25/96 (26%), Positives = 48/96 (49%)
Frame = +2
Query: 19 EELKKKCEQVVVGRNALRQAVKILEKGIENLESENKKLKKDIQEEQAQRKVAIEGKLEIS 78
E +KK+ G+N + K L + I L+S+ KKL+ + + + +Q K +E ++E
Sbjct: 479 EAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEAKGSQAK-TLETEVE-- 649
Query: 79 NTFAALENEVSALISENKKLKQDILEEQAQGKICDQ 114
AL+ + + E +L +D ++Q + DQ
Sbjct: 650 ----ALKKQSEGFLMEYDRLLEDNQSLRSQLQAIDQ 745
Score = 30.8 bits (68), Expect = 2.5
Identities = 25/109 (22%), Positives = 52/109 (46%), Gaps = 12/109 (11%)
Frame = +2
Query: 52 ENKKLKKDIQEEQAQRKVAIEGKLEISNTFAALENEVSALISENKKLKQDILEEQAQGKI 111
E + L+K ++ + Q + +GK S AL E++ L S+ KKL+ + + +Q K
Sbjct: 452 ELRSLRKTMEAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEAKGSQAKT 631
Query: 112 CDQ------------LKKCEKVVEGRNALRQAVKILEKRIENLESENKK 148
+ L + ++++E +LR ++ +++ L +NKK
Sbjct: 632 LETEVEALKKQSEGFLMEYDRLLEDNQSLRSQLQAIDQ--SPLNFDNKK 772
Score = 29.6 bits (65), Expect = 5.7
Identities = 17/62 (27%), Positives = 34/62 (54%), Gaps = 5/62 (8%)
Frame = +2
Query: 595 DELKKKCEKVAVGRNALRQAVKILEKGIENLESENKKLKKE-----NEVSALKSEISALQ 649
+ +KK+ G+N + K L + I L+S+ KKL+ E ++ L++E+ AL+
Sbjct: 479 EAIKKQSRSFEDGKNGNSEEHKALNEEITTLKSKIKKLESECEAKGSQAKTLETEVEALK 658
Query: 650 QK 651
++
Sbjct: 659 KQ 664
>TC229123
Length = 1034
Score = 36.6 bits (83), Expect = 0.046
Identities = 42/179 (23%), Positives = 75/179 (41%), Gaps = 2/179 (1%)
Frame = +3
Query: 635 ENEVSALKSEISALQQKCGAGAREGNG--DVEVLKAGISDTKKEVNRLKKEHVEEERIVA 692
E+E++ K+ L +K G +G D ++ K G T + + E+E+ +
Sbjct: 30 EDELADFKAWFEILSRKSSNGEDQGTSSKDTDLKKKG--STXRGNKPTRNNTAEKEKYSS 203
Query: 693 DSERKTAVDERKNAAAEARKLLEAPKKIAAEVEKQIAKVELRQVHLEKQVNERKMKLAFE 752
+++ DE E KL A E ++ + + + Q N + A +
Sbjct: 204 ENK-----DEFSRLKGEYDKL-------ALEKYSEVTTLLAEKKFMWNQYNIMENDYADK 347
Query: 753 LSKTKEATKRFEAEKKKLLVEKINAESKIKKAQERSESELDKKTADMEKQQAEEQKKLA 811
L +TKEA EK K+LV + K ++ SEL K A+ME + K+++
Sbjct: 348 L-RTKEAEVEKANEKIKILVSSMEQLQSEKYEKDSKISELQSKMAEMEAETKRLNKEIS 521
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.305 0.124 0.307
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,130,462
Number of Sequences: 63676
Number of extensions: 192636
Number of successful extensions: 1408
Number of sequences better than 10.0: 147
Number of HSP's better than 10.0 without gapping: 855
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1361
length of query: 824
length of database: 12,639,632
effective HSP length: 105
effective length of query: 719
effective length of database: 5,953,652
effective search space: 4280675788
effective search space used: 4280675788
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0437b.7


