

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
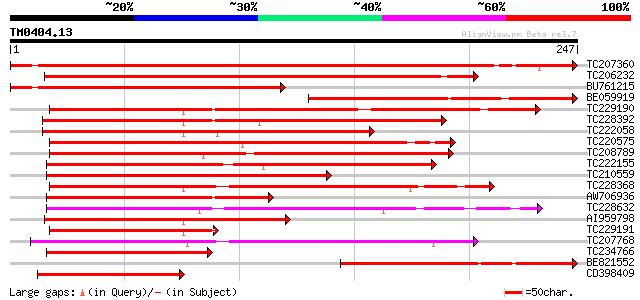
Query= TM0404.13
(247 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207360 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, ... 283 5e-77
TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS... 261 2e-70
BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber... 199 7e-52
BE059919 similar to GP|602900|emb|C SLM1 {Silene latifolia}, par... 158 2e-39
TC229190 similar to UP|Q9SEG4 (Q9SEG4) CAGL2, partial (86%) 156 9e-39
TC228392 similar to UP|Q8LLR0 (Q8LLR0) MADS-box protein 4, parti... 149 1e-36
TC222058 homologue to UP|AGL9_PETHY (Q03489) Agamous-like MADS b... 142 1e-34
TC220575 similar to UP|Q8LLR1 (Q8LLR1) MADS-box protein 3, parti... 139 9e-34
TC208789 similar to UP|Q9SBQ1 (Q9SBQ1) MADS box transcription fa... 137 3e-33
TC222155 weakly similar to UP|O81662 (O81662) Transcription acti... 135 1e-32
TC210559 similar to UP|Q84MI1 (Q84MI1) MADS-box protein (Fragmen... 122 2e-28
TC228368 similar to UP|O49173 (O49173) MADS-box protein NMH 7, p... 120 6e-28
AW706936 115 1e-26
TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, part... 114 4e-26
AI959798 similar to GP|22091481|emb MADS box transcription facto... 112 1e-25
TC229191 homologue to UP|Q84U54 (Q84U54) MADS-RIN-like protein, ... 110 5e-25
TC207768 similar to UP|Q9ST53 (Q9ST53) MADS-box protein 4, parti... 110 5e-25
TC234766 homologue to UP|Q7Y137 (Q7Y137) MADS-box protein PTM5, ... 107 7e-24
BE821552 similar to GP|6970411|dbj| MADS-box protein {Rosa rugos... 104 4e-23
CD398409 103 9e-23
>TC207360 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, partial
(88%)
Length = 1075
Score = 283 bits (724), Expect = 5e-77
Identities = 155/248 (62%), Positives = 188/248 (75%), Gaps = 1/248 (0%)
Frame = +2
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M PNQ+ A S Q+KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 50 MEDPNQAPEA--SSQKKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 223
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
AL+VFS RGRLYEYANNSV+A+IERYKKA + +S S S AN QFYQQE++KLR QI +
Sbjct: 224 ALVVFSTRGRLYEYANNSVRATIERYKKANAAASNAESVSEANTQFYQQESSKLRRQIRD 403
Query: 121 LQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNS 180
+QN NR +LGEAL +++ ++LKNLE +LEKG+SR+RS+K+E LFA++E+MQKREI+L N
Sbjct: 404 IQNLNRHILGEALGSLSLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNH 583
Query: 181 NQLLRAKIAESDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGL-QPNNNQCARQ 239
N LRAKIAE + + NM T ESL Q +D R FF V + + Q +RQ
Sbjct: 584 NNYLRAKIAEHERAQQQQSNMNMSGTLCESL--PSQSYD-RNFFPVNLIASDDQQQYSRQ 754
Query: 240 DQISLQFV 247
D +LQ V
Sbjct: 755 DHTALQLV 778
>TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS-2, partial
(95%)
Length = 960
Score = 261 bits (668), Expect = 2e-70
Identities = 131/189 (69%), Positives = 157/189 (82%)
Frame = +3
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYA 75
R+MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+RGRLYEY+
Sbjct: 69 RRMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYS 248
Query: 76 NNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSN 135
NN+++++IERYKKACSD S +T+ NAQ+YQQE+AKLR QI LQN NR ++G+ALS
Sbjct: 249 NNNIRSTIERYKKACSDHSSASTTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDALST 428
Query: 136 MNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERK 195
+ ++LK LE +LE+GI+RIRSKK+EML AEIEY QKREI+L N N LR KI +D +
Sbjct: 429 LTVKELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKI--TDVER 602
Query: 196 NHNFNMLPG 204
NM+ G
Sbjct: 603 IQQVNMVSG 629
>BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber, partial
(42%)
Length = 450
Score = 199 bits (507), Expect = 7e-52
Identities = 105/120 (87%), Positives = 109/120 (90%)
Frame = +1
Query: 1 MSSPNQSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 60
M+ P+ SMS SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV
Sbjct: 97 MAFPDPSMSV--SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEV 270
Query: 61 ALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISN 120
ALIVFS+RGRLYEYANNSVKA+IERYKKA SDSSG GS S ANAQFYQQEA KLR QISN
Sbjct: 271 ALIVFSSRGRLYEYANNSVKATIERYKKASSDSSGAGSASEANAQFYQQEADKLRQQISN 450
>BE059919 similar to GP|602900|emb|C SLM1 {Silene latifolia}, partial (31%)
Length = 426
Score = 158 bits (399), Expect = 2e-39
Identities = 85/117 (72%), Positives = 93/117 (78%)
Frame = +2
Query: 131 EALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAE 190
E LS MN +DLKNLETKLEKGISRIRSKKNEMLFAEIE+M+KREI LHN NQLLRAKI E
Sbjct: 5 EGLSTMNGKDLKNLETKLEKGISRIRSKKNEMLFAEIEHMKKREIYLHNDNQLLRAKIGE 184
Query: 191 SDERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
ER +HN N L GTT++ES+Q FDSRGFFQVTGLQPNNN +SLQFV
Sbjct: 185 G-ERSHHNVNGLSGTTSYESMQSQ---FDSRGFFQVTGLQPNNNNQYAGQDMSLQFV 343
>TC229190 similar to UP|Q9SEG4 (Q9SEG4) CAGL2, partial (86%)
Length = 1062
Score = 156 bits (394), Expect = 9e-39
Identities = 91/215 (42%), Positives = 130/215 (60%), Gaps = 1/215 (0%)
Frame = +1
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY-AN 76
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FS RG+LYE+ +
Sbjct: 313 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 492
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
NS+ ++ERY+K CS + S G + +E KL+ + +LQ R +LGE L +
Sbjct: 493 NSMLKTLERYQK-CSYGAVEVSKPGKELESSYREYLKLKARFESLQRTQRNLLGEDLGPL 669
Query: 137 NARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKN 196
N +DL+ LE +L+ + K+ + + ++ +Q +E L +N+ L K+ E + R
Sbjct: 670 NTKDLEQLERQLDSSL-----KQTQFMLDQLADLQNKEHMLVEANRSLTMKLEEINSRNQ 834
Query: 197 HNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQP 231
+ G +S+ Q S+GFFQ P
Sbjct: 835 YRQTWEAGE---QSMSYGTQNAHSQGFFQPLECNP 930
>TC228392 similar to UP|Q8LLR0 (Q8LLR0) MADS-box protein 4, partial (98%)
Length = 755
Score = 149 bits (376), Expect = 1e-36
Identities = 80/181 (44%), Positives = 124/181 (68%), Gaps = 5/181 (2%)
Frame = +1
Query: 15 QRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY 74
+R MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FSNRG+ YE+
Sbjct: 31 ERVMGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKQYEF 210
Query: 75 -ANNSVKASIERYKKACSDSSGGGSTSGANAQFY----QQEAAKLRVQISNLQNHNRQML 129
+ +S+ ++ERY+K C+ + + + A QQE +L+ + LQ R ++
Sbjct: 211 CSGSSMLKTLERYQK-CNYGAPEDNVATKEALVLELSSQQEYLRLKARYEALQRSQRNLM 387
Query: 130 GEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIA 189
GE L +++++L++LE +L+ + +IRS + + + ++ +Q++E L SN+ LR ++
Sbjct: 388 GEDLGPLSSKELESLERQLDSSLKQIRSIRTQFMLDQLSDLQRKEHFLGESNRDLRQRLE 567
Query: 190 E 190
E
Sbjct: 568 E 570
>TC222058 homologue to UP|AGL9_PETHY (Q03489) Agamous-like MADS box protein
AGL9 homolog (Floral homeotic protein FBP2) (Floral
binding protein 2), partial (60%)
Length = 495
Score = 142 bits (358), Expect = 1e-34
Identities = 75/147 (51%), Positives = 106/147 (72%), Gaps = 2/147 (1%)
Frame = +3
Query: 15 QRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY 74
+R+MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FSNRG+LYE+
Sbjct: 51 EREMGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEF 230
Query: 75 -ANNSVKASIERYKKA-CSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEA 132
+++S+ ++ERY+K ST A QQE KL+ + LQ R ++GE
Sbjct: 231 CSSSSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGED 410
Query: 133 LSNMNARDLKNLETKLEKGISRIRSKK 159
L +++++L++LE +L+ + +IRS +
Sbjct: 411 LGPLSSKELESLERQLDSSLKQIRSTR 491
>TC220575 similar to UP|Q8LLR1 (Q8LLR1) MADS-box protein 3, partial (44%)
Length = 851
Score = 139 bits (351), Expect = 9e-34
Identities = 75/178 (42%), Positives = 116/178 (65%), Gaps = 1/178 (0%)
Frame = +2
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGRGK+ ++RI+N NRQVTF KRRNGLLKKA+ELSVLCDAE+AL++FS+RG+L++Y++
Sbjct: 158 MGRGKVVLERIQNKINRQVTFSKRRNGLLKKAFELSVLCDAEIALVIFSSRGKLFQYSST 337
Query: 78 SVKASIERYKKACSDSSGGGSTS-GANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
+ IE+Y++ C + S G + + Q QE LRV+ +LQ R +LGE L +
Sbjct: 338 DINRIIEKYRQCCFNMSQTGDVAEHQSEQCLYQELLILRVKHESLQRTQRNLLGEELEPL 517
Query: 137 NARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDER 194
+ ++L +LE +L++ + + R + L + I+ + + +HN Q+ K ES ER
Sbjct: 518 SMKELHSLEKQLDRTLGQARKHLTQKLISRIDELHGK---VHNLEQV--NKHLESQER 676
>TC208789 similar to UP|Q9SBQ1 (Q9SBQ1) MADS box transcription factor,
partial (72%)
Length = 995
Score = 137 bits (346), Expect = 3e-33
Identities = 78/180 (43%), Positives = 109/180 (60%), Gaps = 4/180 (2%)
Frame = +3
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGRG++E+KRIEN QVTF KRR+GLLKKA E+SVLCDA+VALIVFS +G+L +Y+N
Sbjct: 219 MGRGRVELKRIENKIFMQVTFSKRRSGLLKKAREISVLCDADVALIVFSTKGKLLDYSNQ 398
Query: 78 SVKASI----ERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEAL 133
I ERY A G N + E KL+ ++ LQ + R +GE L
Sbjct: 399 PCTERILERYERYSYAERQLVGDDQPPNEN---WVIEHEKLKARVEVLQRNQRNFMGEDL 569
Query: 134 SNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDE 193
++N R L++LE +L+ + IRS+KN+ + I +QK++ L N LL KI E ++
Sbjct: 570 DSLNLRGLQSLEQQLDSALKHIRSRKNQAMNESISELQKKDRTLREHNNLLSKKIKEKEK 749
>TC222155 weakly similar to UP|O81662 (O81662) Transcription activator,
partial (65%)
Length = 915
Score = 135 bits (341), Expect = 1e-32
Identities = 71/173 (41%), Positives = 116/173 (67%), Gaps = 3/173 (1%)
Frame = +2
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+M RGK+++K+IE+TT+RQV F KRR+GLLKKAYELSVLCDAEVA+IVFS GRLYE+++
Sbjct: 149 EMARGKVQLKKIEDTTSRQVAFSKRRSGLLKKAYELSVLCDAEVAVIVFSQNGRLYEFSS 328
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQ---EAAKLRVQISNLQNHNRQMLGEAL 133
+ + +ERY++ D S + QQ ++A L +I L++ R++LG+++
Sbjct: 329 SDMTKILERYREHTKDV----PASKFGDDYIQQLKLDSASLAKKIELLEHSKRELLGQSV 496
Query: 134 SNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRA 186
S+ + +LK +E +L+ + R+R +K ++ +I+ ++ +E +L N L A
Sbjct: 497 SSCSYDELKGIEEQLQISLQRVRQRKTQLYTEQIDQLRSQESNLLKENAKLSA 655
>TC210559 similar to UP|Q84MI1 (Q84MI1) MADS-box protein (Fragment), partial
(64%)
Length = 710
Score = 122 bits (306), Expect = 2e-28
Identities = 62/124 (50%), Positives = 89/124 (71%)
Frame = +1
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+M RG+ +++RIEN T+RQVTF KRRNGLLKKA+ELSVLCDAEVALI+FS RG+LYE+A+
Sbjct: 337 EMVRGETQMRRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFAS 516
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNM 136
+S++ +IERY++ + + N Q +QE A L +I L+ R++LGE L +
Sbjct: 517 SSMQDTIERYRRHNRSAQTVNRSDEQNMQHLKQETANLMKKIELLEASKRKLLGEGLGSC 696
Query: 137 NARD 140
+ +
Sbjct: 697 SLEE 708
>TC228368 similar to UP|O49173 (O49173) MADS-box protein NMH 7, partial (97%)
Length = 1042
Score = 120 bits (301), Expect = 6e-28
Identities = 73/199 (36%), Positives = 121/199 (60%), Gaps = 5/199 (2%)
Frame = +3
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY--A 75
M RGKI+IKRIEN TNRQVT+ KRRNGL KKA EL+VLCDA+V++I+FS+ G+L++Y
Sbjct: 81 MARGKIQIKRIENNTNRQVTYSKRRNGLFKKANELTVLCDAKVSIIMFSSTGKLHQYISP 260
Query: 76 NNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSN 135
+ S K ++Y+ G ++ + Q+ KL+ NL+ RQ +G+ L+
Sbjct: 261 STSTKQFFDQYQMTL-----GVDLWNSHYENMQENLKKLKEVNRNLRKEIRQRMGDCLNE 425
Query: 136 MNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKR---EIDLHNSNQLLRAKIAESD 192
+ DLK LE +++K +R +K +++ +I+ +K+ E ++H N+LLR A ++
Sbjct: 426 LGMEDLKLLEEEMDKAAKVVRERKYKVITNQIDTQRKKFNNEKEVH--NRLLRDLDARAE 599
Query: 193 ERKNHNFNMLPGTTNFESL 211
+ + F ++ +ES+
Sbjct: 600 DPR---FALIDNGGEYESV 647
>AW706936
Length = 422
Score = 115 bits (289), Expect = 1e-26
Identities = 60/99 (60%), Positives = 76/99 (76%)
Frame = +2
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
KMGRGKI I+RI+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV L+VFS+ G+LY+YA+
Sbjct: 128 KMGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKARELSILCDAEVGLMVFSSTGKLYDYAS 307
Query: 77 NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLR 115
S+K+ IERY K + + A+F+Q EAA LR
Sbjct: 308 TSMKSVIERYNK-LKEEHHHLMNPASEAKFWQTEAASLR 421
>TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, partial (82%)
Length = 880
Score = 114 bits (285), Expect = 4e-26
Identities = 77/221 (34%), Positives = 125/221 (55%), Gaps = 5/221 (2%)
Frame = +1
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+MGRGKIEIKRIEN++NRQVT+ KR+NG+LKKA E++VLCDA+V+LI+F+ G++++Y +
Sbjct: 52 EMGRGKIEIKRIENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYIS 231
Query: 77 NSVKA--SIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALS 134
S +ERY K + G A + E +L+ + ++Q R + G+ ++
Sbjct: 232 PSTTLIDILERYHK-----TSGKRLWDAKHENLNGEIERLKKENDSMQIELRHLKGDDIN 396
Query: 135 NMNARDLKNLETKLEKGISRIRSKKNE---MLFAEIEYMQKREIDLHNSNQLLRAKIAES 191
++N ++L LE LE G+ +R K+ + ML + +++ +L N L + ++AE
Sbjct: 397 SLNYKELMALEDALETGLVSVREKQMDVYRMLRRNDKILEEENREL---NFLWQQRLAEE 567
Query: 192 DERKNHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPN 232
R+ N G S PF F+V +QPN
Sbjct: 568 GAREVDN-----GFDQSVRDYNSHMPF----AFRVQPMQPN 663
>AI959798 similar to GP|22091481|emb MADS box transcription factor {Daucus
carota subsp. sativus}, partial (39%)
Length = 409
Score = 112 bits (281), Expect = 1e-25
Identities = 60/108 (55%), Positives = 75/108 (68%), Gaps = 1/108 (0%)
Frame = +3
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY- 74
+ MGRGK+E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FSNRG+LYE+
Sbjct: 84 KHMGRGKVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFC 263
Query: 75 ANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQ 122
+ +S ++ERY + + Q QE KL+ ++ LQ
Sbjct: 264 SGHSTAKTLERYHRCSYGALEVQHQPEIETQRRYQEYLKLKSRVEALQ 407
>TC229191 homologue to UP|Q84U54 (Q84U54) MADS-RIN-like protein, partial
(32%)
Length = 558
Score = 110 bits (276), Expect = 5e-25
Identities = 55/75 (73%), Positives = 68/75 (90%), Gaps = 1/75 (1%)
Frame = +2
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY-AN 76
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FSNRG+LYE+ ++
Sbjct: 320 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 499
Query: 77 NSVKASIERYKKACS 91
+S+ ++ERY+K CS
Sbjct: 500 SSMLKTLERYQK-CS 541
>TC207768 similar to UP|Q9ST53 (Q9ST53) MADS-box protein 4, partial (27%)
Length = 951
Score = 110 bits (276), Expect = 5e-25
Identities = 70/198 (35%), Positives = 109/198 (54%), Gaps = 3/198 (1%)
Frame = +3
Query: 10 ANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRG 69
A S +R+MG+ K+EIKRIEN + RQ+TF KRRNGL+KKA ELS+LCDA+VAL++FS+ G
Sbjct: 87 ARVSARRRMGKKKVEIKRIENKSTRQITFSKRRNGLMKKARELSILCDAKVALLIFSSTG 266
Query: 70 RLYEYAN-NSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQM 128
+LYE N +S+ +++Y G S + +Q E A + + Q R
Sbjct: 267 KLYELCNGDSLAEVVQQYWDHL-----GASGTDTKSQELCFEIADIWSGSAFSQMIKRHF 431
Query: 129 LGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQL--LRA 186
L +++ DL LE +SRIRS K ++ + ++K+ L +N + +
Sbjct: 432 GVSELEHLSVSDLMELEKLTHAALSRIRSAKMRLMMESVVNLKKKIEALEKTNDVNNVVT 611
Query: 187 KIAESDERKNHNFNMLPG 204
+ + D+ N+ PG
Sbjct: 612 RSIDCDQTDGVTHNLFPG 665
>TC234766 homologue to UP|Q7Y137 (Q7Y137) MADS-box protein PTM5, partial
(33%)
Length = 490
Score = 107 bits (266), Expect = 7e-24
Identities = 50/72 (69%), Positives = 66/72 (91%)
Frame = +2
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+M RGK +++RIEN T+RQVTF KRRNGLLKKA+ELSVLCDAEVALI+FS RG+LYE+A+
Sbjct: 236 EMVRGKTQLRRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFAS 415
Query: 77 NSVKASIERYKK 88
+S++ +IERY++
Sbjct: 416 SSMQDTIERYRR 451
>BE821552 similar to GP|6970411|dbj| MADS-box protein {Rosa rugosa}, partial
(29%)
Length = 544
Score = 104 bits (259), Expect = 4e-23
Identities = 55/103 (53%), Positives = 74/103 (71%)
Frame = -2
Query: 145 ETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHNFNMLPG 204
E++LEKG+SR+RS+K+E LFA+IE+MQKREI+L N N LRAKIAE++ + +M+PG
Sbjct: 543 ESRLEKGLSRVRSRKHETLFADIEFMQKREIELQNHNNFLRAKIAENERAQQRQQDMIPG 364
Query: 205 TTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
T ES + Q +D R FF V + NNNQ +RQDQ +LQ V
Sbjct: 363 -TECESTIPNSQSYD-RNFFPVNLIDSNNNQYSRQDQTALQLV 241
>CD398409
Length = 639
Score = 103 bits (256), Expect = 9e-23
Identities = 48/64 (75%), Positives = 57/64 (89%)
Frame = -1
Query: 13 SPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLY 72
+P + MGRGKI I+RIEN+TNRQVTFCKRRNGLLKK ELS+LCDAEV +IVFS+ G+LY
Sbjct: 564 NPSK*MGRGKIPIRRIENSTNRQVTFCKRRNGLLKKTRELSILCDAEVGVIVFSSTGKLY 385
Query: 73 EYAN 76
EY+N
Sbjct: 384 EYSN 373
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,206,165
Number of Sequences: 63676
Number of extensions: 87102
Number of successful extensions: 403
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 395
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 395
length of query: 247
length of database: 12,639,632
effective HSP length: 95
effective length of query: 152
effective length of database: 6,590,412
effective search space: 1001742624
effective search space used: 1001742624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0404.13


