

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.11
(365 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
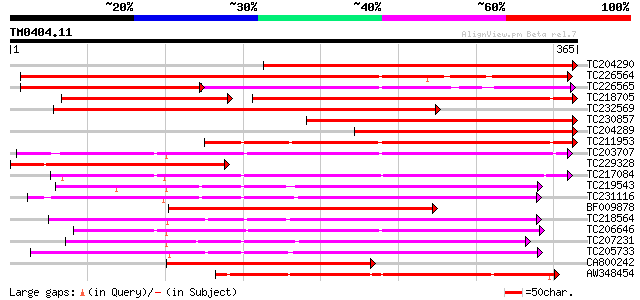
Score E
Sequences producing significant alignments: (bits) Value
TC204290 371 e-103
TC226564 308 2e-84
TC226565 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif lipase/... 189 2e-83
TC218705 weakly similar to GB|AAO63389.1|28950931|BT005325 At1g7... 196 7e-79
TC232569 288 3e-78
TC230857 280 7e-76
TC204289 257 5e-69
TC211953 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/... 253 7e-68
TC203707 245 3e-65
TC229328 235 3e-62
TC217084 235 3e-62
TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase E... 219 2e-57
TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 216 2e-56
BF009878 215 2e-56
TC218564 204 7e-53
TC206646 200 1e-51
TC207231 similar to UP|Q94CH6 (Q94CH6) Family II lipase EXL3, pa... 199 2e-51
TC205733 197 6e-51
CA800242 179 2e-45
AW348454 similar to GP|21537293|gb GDSL-motif lipase/hydrolase-l... 175 3e-44
>TC204290
Length = 823
Score = 371 bits (952), Expect = e-103
Identities = 173/202 (85%), Positives = 190/202 (93%)
Frame = +2
Query: 164 SIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQ 223
SIGLGSNDYLNNYFMPQFYS+SRQY+ D+YADVLIQAYTEQL+TLYN+GARKMVLFGIGQ
Sbjct: 2 SIGLGSNDYLNNYFMPQFYSSSRQYSPDEYADVLIQAYTEQLKTLYNYGARKMVLFGIGQ 181
Query: 224 IGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQD 283
IGCSPNELAQ S DG+TCV IN ANQIFNNKLK + DQFNNQLPDA+VIYIN+YGIFQD
Sbjct: 182 IGCSPNELAQNSPDGKTCVEKINTANQIFNNKLKGLTDQFNNQLPDAKVIYINSYGIFQD 361
Query: 284 IIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQ 343
II++P+ YGFS TN GCCGVGRNNGQITCLPMQTPC+NRREYLFWDAFHP+EAGNVV+AQ
Sbjct: 362 IISNPSAYGFSVTNAGCCGVGRNNGQITCLPMQTPCQNRREYLFWDAFHPTEAGNVVVAQ 541
Query: 344 RAYSAQSPSDAYPIDIKRLAQI 365
RAYSAQS SDAYP+DI+RLAQI
Sbjct: 542 RAYSAQSASDAYPVDIQRLAQI 607
>TC226564
Length = 1337
Score = 308 bits (790), Expect = 2e-84
Identities = 157/358 (43%), Positives = 226/358 (62%), Gaps = 3/358 (0%)
Frame = +3
Query: 8 MVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG 67
+V+ ++ +V V QVPC FIFGDS+ D+GNNN L + +++++ PYGIDFP G
Sbjct: 69 LVMFLVFLVANCMQHCVHGVSQVPCLFIFGDSMSDSGNNNELPTTSKSNFRPYGIDFPLG 248
Query: 68 PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGR 127
P+GR++NG+T +D+I + LGF+ FIPP+ +TSG DIL+GVN+AS +GIR ETG G
Sbjct: 249 PTGRYTNGRTEIDIITQFLGFEKFIPPFANTSGSDILKGVNYASGGSGIRNETGWHYGAA 428
Query: 128 ISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQ 187
I Q+ N++ VS++ LG+ D A YL KC+Y + +GSNDY+ NYF+P FY TS
Sbjct: 429 IGLGLQLANHRVIVSEIATKLGSPDLARQYLEKCLYYVNIGSNDYMGNYFLPPFYPTSTI 608
Query: 188 YTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDIND 247
YT +++ VLI+ + LQ L++ GARK L G+G IGC+P ++ +G +C + N
Sbjct: 609 YTIEEFTQVLIEELSLNLQALHDIGARKYALAGLGLIGCTPGMVSAHGTNG-SCAEEQNL 785
Query: 248 ANQIFNNKLKSVVDQFNNQL--PDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGR 305
A FNNKLK+ VDQFNN +++ I+IN + I YGF CC G
Sbjct: 786 AAFNFNNKLKARVDQFNNDFYYANSKFIFINTQAL---AIELRDKYGFPVPETPCCLPGL 956
Query: 306 NNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSD-AYPIDIKRL 362
C+P Q PC NR +Y+F+DAFHP+E N++ A +Y++ + S YP+DIK L
Sbjct: 957 TG---ECVPDQEPCYNRNDYVFFDAFHPTEQWNLLNALTSYNSTTNSAFTYPMDIKHL 1121
>TC226565 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif
lipase/hydrolase-like protein, partial (25%)
Length = 1448
Score = 189 bits (481), Expect(2) = 2e-83
Identities = 104/243 (42%), Positives = 147/243 (59%), Gaps = 2/243 (0%)
Frame = +3
Query: 124 LGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYS 183
LG ISF Q+ N++ VSQ+ + LG+ D A YL KC+Y + +GSNDY+NNYF+PQ Y
Sbjct: 510 LGATISFGLQLANHRVIVSQIASRLGSSDLALQYLEKCLYYVNIGSNDYMNNYFLPQLYP 689
Query: 184 TSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVS 243
SR Y+ +QYA LI+ + L L++ GARK VL +G+IGC+P+ + +G +CV
Sbjct: 690 ASRIYSLEQYAQALIEELSLNLLALHDLGARKYVLARLGRIGCTPSVMHSHGTNG-SCVE 866
Query: 244 DINDANQIFNNKLKSVVDQFNNQL-PDARVIYINAYGIFQDIIASPATYGFSNTNEGCCG 302
+ N A +NNKLK++VDQFN++ +++ I I DI +GF ++ CC
Sbjct: 867 EQNAATSDYNNKLKALVDQFNDRFSANSKFILIPNESNAIDI-----AHGFLVSDAACCP 1031
Query: 303 VGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAY-SAQSPSDAYPIDIKR 361
G C P Q PC NR +YLFWD HP+EA N+V A Y S P+ YP+DIK+
Sbjct: 1032SG-------CNPDQKPCNNRSDYLFWDEVHPTEAWNLVNAISVYNSTIGPAFNYPMDIKQ 1190
Query: 362 LAQ 364
L +
Sbjct: 1191LVE 1199
Score = 137 bits (346), Expect(2) = 2e-83
Identities = 64/118 (54%), Positives = 85/118 (71%)
Frame = +2
Query: 8 MVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG 67
+V+ +L + V QVPC FIFGDSL D+GNNN L + A+++Y PYGIDFP G
Sbjct: 161 LVMVLLFLAANYLQDCVHGVSQVPCLFIFGDSLSDSGNNNELPTSAKSNYRPYGIDFPLG 340
Query: 68 PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLG 125
P+GRF+NG+T +D+I +LLGF+ FIPP+ +TSG +IL+GVN+AS AGIR ET G
Sbjct: 341 PTGRFTNGRTEIDIITQLLGFEKFIPPFANTSGSNILKGVNYASGGAGIRVETQLAFG 514
>TC218705 weakly similar to GB|AAO63389.1|28950931|BT005325 At1g71250
{Arabidopsis thaliana;} , partial (78%)
Length = 1324
Score = 196 bits (498), Expect(2) = 7e-79
Identities = 89/209 (42%), Positives = 131/209 (62%)
Frame = +1
Query: 157 YLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKM 216
+L+K I + GSNDY+NNY +P Y +SR YT + ++L+ +Y Q+ L++ G RK
Sbjct: 520 FLAKSIAVVVTGSNDYINNYLLPGLYGSSRNYTAQDFGNLLVNSYVRQILALHSVGLRKF 699
Query: 217 VLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYIN 276
L GIG +GC P A CV +N FN L+S+VDQ N P+A +Y N
Sbjct: 700 FLAGIGPLGCIPTLRAAALAPTGRCVDLVNQMVGTFNEGLRSMVDQLNRNHPNAIFVYGN 879
Query: 277 AYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEA 336
Y +F DI+ +PA + F+ + CCG+GRN GQ+TCLP+Q PC +R +Y+FWDAFHP+E+
Sbjct: 880 TYRVFGDILNNPAAFAFNVVDRACCGIGRNRGQLTCLPLQFPCTSRNQYVFWDAFHPTES 1059
Query: 337 GNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
V A R + +P D+YPI+++++A I
Sbjct: 1060ATYVFAWRVVNG-APDDSYPINMQQMATI 1143
Score = 116 bits (290), Expect(2) = 7e-79
Identities = 60/111 (54%), Positives = 79/111 (71%), Gaps = 1/111 (0%)
Frame = +2
Query: 34 FIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIP 93
F+FGDSLV+ GNNN L ++ARA+Y PYGIDF +GRFSNGK+ +D I +LLG P
Sbjct: 149 FVFGDSLVEVGNNNFLNTIARANYFPYGIDFSKSSTGRFSNGKSLIDFIGDLLGIPSPTP 328
Query: 94 -PYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQ 143
ST G IL GVN+ASA+AGI +E+G+ G R S S QV N+++T++Q
Sbjct: 329 FADPSTVGTRILYGVNYASASAGILDESGRHYGDRYSLSQQVLNFENTLNQ 481
>TC232569
Length = 974
Score = 288 bits (736), Expect = 3e-78
Identities = 126/249 (50%), Positives = 185/249 (73%)
Frame = +2
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
QVPC F+ GDSL DNGNNN L++ A ++Y PYGID+P GP+GRF+NGK +D I+E LGF
Sbjct: 185 QVPCTFVLGDSLSDNGNNNNLQTNASSNYRPYGIDYPTGPTGRFTNGKNIIDFISEYLGF 364
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
+ IPP +TSG DIL+G N+AS AAGI ++G+ LG I Q++N+++T++++V L
Sbjct: 365 TEPIPPNANTSGSDILKGANYASGAAGIPFKSGKHLGDNIHLGEQIRNHRATITKIVRRL 544
Query: 149 GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTL 208
G +A YL KC+Y + +GSNDY+NNYF+PQFY TSR YT ++Y D+LI+ Y++ ++ L
Sbjct: 545 GGSGRAREYLKKCLYYVNIGSNDYINNYFLPQFYPTSRTYTLERYTDILIKQYSDDIKKL 724
Query: 209 YNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLP 268
+ GARK + G+G IGC+PN +++R +G CV+++N+A +F+NKLKS VDQF N P
Sbjct: 725 HRSGARKFAIVGLGLIGCTPNAISRRGTNGEVCVAELNNAAFLFSNKLKSQVDQFKNTFP 904
Query: 269 DARVIYINA 277
D++ ++N+
Sbjct: 905 DSKFSFVNS 931
>TC230857
Length = 768
Score = 280 bits (716), Expect = 7e-76
Identities = 126/174 (72%), Positives = 149/174 (85%)
Frame = +2
Query: 192 QYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQI 251
QYADVL+QAY +QL+ LY +GARKM LFG+GQIGCSPN LAQ S DGRTCV+ IN ANQ+
Sbjct: 5 QYADVLVQAYAQQLRILYKYGARKMALFGVGQIGCSPNALAQNSPDGRTCVARINSANQL 184
Query: 252 FNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQIT 311
FNN L+S+VDQ NNQ+PDAR IYIN YGIFQDI+++P++YGF TN GCCGVGRNNGQ+T
Sbjct: 185 FNNGLRSLVDQLNNQVPDARFIYINVYGIFQDILSNPSSYGFRVTNAGCCGVGRNNGQVT 364
Query: 312 CLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
CLP+QTPC R +LFWDAFHP+EA N +I +RAY+AQS SDAYP+DI RLAQI
Sbjct: 365 CLPLQTPCRTRGAFLFWDAFHPTEAANTIIGRRAYNAQSASDAYPVDINRLAQI 526
>TC204289
Length = 635
Score = 257 bits (657), Expect = 5e-69
Identities = 118/143 (82%), Positives = 130/143 (90%)
Frame = +2
Query: 223 QIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQ 282
QIGCSP LAQ S DG+TCV IN ANQIFNNKLK + DQFNNQLPDARVIY+N+YGIFQ
Sbjct: 2 QIGCSPXXLAQNSPDGKTCVEKINSANQIFNNKLKGLTDQFNNQLPDARVIYVNSYGIFQ 181
Query: 283 DIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIA 342
DII++P+ YGFS TN GCCGVGRNNGQITCLPMQTPC+NRREYLFWDAFHP+EAGNVV+A
Sbjct: 182 DIISNPSAYGFSVTNAGCCGVGRNNGQITCLPMQTPCQNRREYLFWDAFHPTEAGNVVVA 361
Query: 343 QRAYSAQSPSDAYPIDIKRLAQI 365
QRAYSAQS SDAYP+DI+RLAQI
Sbjct: 362 QRAYSAQSASDAYPVDIQRLAQI 430
>TC211953 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (37%)
Length = 1023
Score = 253 bits (647), Expect = 7e-68
Identities = 118/240 (49%), Positives = 171/240 (71%)
Frame = +1
Query: 126 GRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTS 185
GRI F Q+ N+++T++Q+ LG D L++CI+ +G+GSNDYLNNY MP Y T
Sbjct: 214 GRIPFDQQLSNFENTLNQITGNLG-ADYMGTALARCIFFVGMGSNDYLNNYLMPN-YPTR 387
Query: 186 RQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDI 245
QY QYAD+L+Q Y++QL LYN GARK V+ G+G++GC P+ LAQ + TC ++
Sbjct: 388 NQYNGQQYADLLVQTYSQQLTRLYNLGARKFVIAGLGEMGCIPSILAQSTTG--TCSEEV 561
Query: 246 NDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGR 305
N Q FN +K+++ FNN LP AR I+ ++ +FQDI+ + +YGF+ N GCCG+GR
Sbjct: 562 NLLVQPFNENVKTMLGNFNNNLPGARFIFADSSRMFQDILLNARSYGFAVVNRGCCGIGR 741
Query: 306 NNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
N GQITCLP QTPC NRR+Y+FWDAFHP+EA N+++ + A++ +P+ YPI+I++LA++
Sbjct: 742 NRGQITCLPFQTPCPNRRQYVFWDAFHPTEAVNILMGRMAFNG-NPNFVYPINIRQLAEL 918
>TC203707
Length = 1394
Score = 245 bits (625), Expect = 3e-65
Identities = 135/361 (37%), Positives = 206/361 (56%), Gaps = 3/361 (0%)
Frame = +1
Query: 5 VNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDF 64
+N++++ ++++V+ L G A +F+FGDSLVDNGNNN L + ARAD PYGID+
Sbjct: 43 INIVLLGIVLVVVRLKGAEAQRA-----FFVFGDSLVDNGNNNFLATTARADAPPYGIDY 207
Query: 65 PGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTS--GDDILRGVNFASAAAGIREETG 121
P G P+GRFSNG D I++ LG + +P Y+ G+ +L G NFASA GI +TG
Sbjct: 208 PTGRPTGRFSNGYNIPDFISQSLGAESTLP-YLDPELDGERLLVGANFASAGIGILNDTG 384
Query: 122 QQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQF 181
Q I Q++ ++ +V ++G E Q ++ + I LG ND++NNY++ +
Sbjct: 385 IQFVNIIRIYRQLEYWEEYQQRVSGLIGPE-QTERLINGALVLITLGGNDFVNNYYLVPY 561
Query: 182 YSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTC 241
+ SRQY Y +I Y + L+ LY GAR++++ G G +GC P ELAQRS +G C
Sbjct: 562 SARSRQYNLPDYVKYIISEYKKVLRRLYEIGARRVLVTGTGPLGCVPAELAQRSTNG-DC 738
Query: 242 VSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCC 301
+++ A +FN +L ++ Q N+++ + +N + D I++P YGF + CC
Sbjct: 739 SAELQQAAALFNPQLVQIIRQLNSEIGSNVFVGVNTQQMHIDFISNPQRYGFVTSKVACC 918
Query: 302 GVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKR 361
G G NG C P C NR Y FWD FHP+E N +I Q+ S S YP+++
Sbjct: 919 GQGPYNGLGLCTPASNLCPNRDSYAFWDPFHPTERANRIIVQQILSGTS-EYMYPMNLST 1095
Query: 362 L 362
+
Sbjct: 1096I 1098
>TC229328
Length = 514
Score = 235 bits (599), Expect = 3e-62
Identities = 113/141 (80%), Positives = 126/141 (89%)
Frame = +2
Query: 1 MDLNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPY 60
+DL ++M+ + ++V+ LGLWGG V APQVPCYFIFGDSLVDNGNNN L+SLARADY+PY
Sbjct: 95 LDLTISMLALIVVVVSLGLWGG-VQGAPQVPCYFIFGDSLVDNGNNNQLQSLARADYLPY 271
Query: 61 GIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREET 120
GIDFPGGPSGRFSNGKTTVD IAELLGFDD+IPPY SGD IL+GVN+ASAAAGIREET
Sbjct: 272 GIDFPGGPSGRFSNGKTTVDAIAELLGFDDYIPPYADASGDAILKGVNYASAAAGIREET 451
Query: 121 GQQLGGRISFSGQVQNYQSTV 141
GQQLGGRI FSGQV NYQSTV
Sbjct: 452 GQQLGGRIIFSGQVHNYQSTV 514
>TC217084
Length = 1310
Score = 235 bits (599), Expect = 3e-62
Identities = 133/342 (38%), Positives = 193/342 (55%), Gaps = 6/342 (1%)
Frame = +3
Query: 27 APQVPC---YFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVI 82
APQ +F+FGDSLVDNGNNN L + ARAD PYGID SGRFSNG D+I
Sbjct: 141 APQAEAARAFFVFGDSLVDNGNNNFLATTARADSYPYGIDSASHRASGRFSNGLNMPDLI 320
Query: 83 AELLGFDDFIPPYVST--SGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQST 140
+E +G + +P Y+S +G+ +L G NFASA GI +TG Q I + Q+ ++
Sbjct: 321 SEKIGSEPTLP-YLSPQLNGERLLVGANFASAGIGILNDTGIQFINIIRITEQLAYFKQY 497
Query: 141 VSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQA 200
+V ++G E+Q N ++K + I LG ND++NNY++ F + SR+Y Y LI
Sbjct: 498 QQRVSALIG-EEQTRNLVNKALVLITLGGNDFVNNYYLVPFSARSREYALPDYVVFLISE 674
Query: 201 YTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVV 260
Y + L LY GAR++++ G G +GC P ELA SQ+G C +++ A +FN +L ++
Sbjct: 675 YRKILANLYELGARRVLVTGTGPLGCVPAELAMHSQNGE-CATELQRAVNLFNPQLVQLL 851
Query: 261 DQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCE 320
+ N Q+ I NA+ + D +++P YGF + CCG G NG C P C
Sbjct: 852 HELNTQIGSDVFISANAFTMHLDFVSNPQAYGFVTSKVACCGQGAYNGIGLCTPASNLCP 1031
Query: 321 NRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRL 362
NR Y FWD FHPSE N +I + + S +P+++ +
Sbjct: 1032NRDLYAFWDPFHPSERANRLIVDK-FMTGSTEYMHPMNLSTI 1154
>TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase EXL1, partial
(49%)
Length = 1085
Score = 219 bits (557), Expect = 2e-57
Identities = 119/320 (37%), Positives = 180/320 (56%), Gaps = 6/320 (1%)
Frame = +2
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG--PSGRFSNGKTTVDVIAELLG 87
+P +FGDS+VD GNNN + ++A+ +++PYG DF GG P+GRFSNG T D+IA G
Sbjct: 116 IPAVIVFGDSIVDTGNNNYITTIAKCNFLPYGRDFGGGNQPTGRFSNGLTPSDIIAAKFG 295
Query: 88 FDDFIPPYVSTS--GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVV 145
+ +PPY+ D+L GV+FAS A+G T + + +S S Q+ ++ ++++
Sbjct: 296 VKELLPPYLDPKLQPQDLLTGVSFASGASGYDPLTSK-IASALSLSDQLDTFREYKNKIM 472
Query: 146 NILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQL 205
I+G E++ A +SK IY + GSND N YF+ +Y Y D++ T L
Sbjct: 473 EIVG-ENRTATIISKSIYILCTGSNDITNTYFV-----RGGEYDIQAYTDLMASQATNFL 634
Query: 206 QTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNN 265
Q LY GAR++ + G+ +GC P++ R C N+A +FN+KL S +D
Sbjct: 635 QELYGLGARRIGVVGLPVLGCVPSQRTLHGGIFRACSDFENEAAVLFNSKLSSQMDALKK 814
Query: 266 QLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQ-TPCENRRE 324
Q +AR +Y++ Y ++I +PA YGF ++GCCG G+ C C N
Sbjct: 815 QFQEARFVYLDLYNPVLNLIQNPAKYGFEVMDQGCCGTGKLEVGPLCNHFTLLICSNTSN 994
Query: 325 YLFWDAFHPSEAG-NVVIAQ 343
Y+FWD+FHP+EA NVV Q
Sbjct: 995 YIFWDSFHPTEAAYNVVCTQ 1054
>TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (94%)
Length = 1541
Score = 216 bits (549), Expect = 2e-56
Identities = 121/335 (36%), Positives = 183/335 (54%), Gaps = 4/335 (1%)
Frame = +1
Query: 12 MLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSG 70
+L ++L + G P VP F FGDS+VD GNNN ++ +A++ PYG DF P+G
Sbjct: 397 LLAVLLNVTNG----QPLVPAIFTFGDSIVDVGNNNHQLTIVKANFPPYGRDFENHFPTG 564
Query: 71 RFSNGKTTVDVIAELLGFDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRI 128
RF NGK D IA++LGF + P Y++ T G ++L G NFASA++G E T + L I
Sbjct: 565 RFCNGKLATDFIADILGFTSYQPAYLNLKTKGKNLLNGANFASASSGYFELTSK-LYSSI 741
Query: 129 SFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQY 188
S Q++ Y+ +++V G + A++ +S IY I G++D++ NY++ + + Y
Sbjct: 742 PLSKQLEYYKECQTKLVEAAG-QSSASSIISDAIYLISAGTSDFVQNYYINPLLN--KLY 912
Query: 189 TTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDA 248
TTDQ++D L++ Y+ +Q+LY GAR++ + + IGC P + CV+ +N
Sbjct: 913 TTDQFSDTLLRCYSNFIQSLYALGARRIGVTSLPPIGCLPAVITLFGAHINECVTSLNSD 1092
Query: 249 NQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNG 308
FN KL + N LP ++ + Y D+ P+ GF + CCG G
Sbjct: 1093AINFNEKLNTTSQNLKNMLPGLNLVVFDIYQPLYDLATKPSENGFFEARKACCGTGLIEV 1272
Query: 309 QITCLPMQT-PCENRREYLFWDAFHPSEAGNVVIA 342
I C C N EY+FWD FHPSEA N V+A
Sbjct: 1273SILCNKKSIGTCANASEYVFWDGFHPSEAANKVLA 1377
>BF009878
Length = 520
Score = 215 bits (548), Expect = 2e-56
Identities = 108/173 (62%), Positives = 131/173 (75%)
Frame = +2
Query: 103 ILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCI 162
IL GVN+ASAAAGIREETGQ+LGG+ISF+GQVQNYQ TVSQ+VN+LG+E+ ANYL KCI
Sbjct: 2 ILSGVNYASAAAGIREETGQRLGGQISFTGQVQNYQRTVSQMVNLLGDENTTANYLIKCI 181
Query: 163 YSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIG 222
YSI +GSN+YLNNYFMP YS S Q+T Y +VLIQAYT+QL+ LY + RK+ LF I
Sbjct: 182 YSIEMGSNNYLNNYFMPLIYSISIQFTPQHYTNVLIQAYTQQLKILYKYNTRKIALFSIN 361
Query: 223 QIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYI 275
QI SPN L+Q RT ++ IN NQ+FNN L S++ Q NN P R IY+
Sbjct: 362 QIDYSPNTLSQNMPXDRTYIAIINAGNQLFNNILISLIYQLNNHCPYTRFIYM 520
>TC218564
Length = 1398
Score = 204 bits (518), Expect = 7e-53
Identities = 113/321 (35%), Positives = 170/321 (52%), Gaps = 4/321 (1%)
Frame = +1
Query: 26 AAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAE 84
A VP +FGDS VD+GNNN + ++ ++++ PYG DF G P+GRF NG+ D IAE
Sbjct: 73 AKNNVPAVIVFGDSSVDSGNNNVIATVLKSNFKPYGRDFEGDRPTGRFCNGRVPPDFIAE 252
Query: 85 LLGFDDFIPPYVSTSG--DDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVS 142
G +P Y+ + D GV FASA G T L I +++ Y+ +
Sbjct: 253 AFGIKRTVPAYLDPAYTIQDFATGVCFASAGTGYDNATSAVLNV-IPLWKEIEYYKEYQA 429
Query: 143 QVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYT 202
++ LG E +A +S+ +Y + LG+ND+L NY++ F + +T QY D L++
Sbjct: 430 KLRTHLGVE-KANKIISEALYLMSLGTNDFLENYYV--FPTRRLHFTVSQYQDFLLRIAE 600
Query: 203 EQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQ 262
++ LY G RK+ + G+G +GC P E A C + ND FN KL++V+ +
Sbjct: 601 NFVRELYALGVRKLSITGLGPVGCLPLERATNILGDHGCNQEYNDVALSFNRKLENVITK 780
Query: 263 FNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQ-TPCEN 321
N +LP + + NAY I DII P+TYGF + CC G C C +
Sbjct: 781 LNRELPRLKALSANAYSIVNDIITKPSTYGFEVVEKACCSTGTFEMSYLCSDKNPLTCTD 960
Query: 322 RREYLFWDAFHPSEAGNVVIA 342
+Y+FWDAFHP+E N +++
Sbjct: 961 AEKYVFWDAFHPTEKTNRIVS 1023
>TC206646
Length = 1178
Score = 200 bits (508), Expect = 1e-51
Identities = 116/306 (37%), Positives = 170/306 (54%), Gaps = 3/306 (0%)
Frame = +1
Query: 42 DNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTS- 99
D+GNN+ L + ARAD PYGID+P P+GRFSNG D+I+ LG +P Y+S
Sbjct: 7 DSGNNDFLVTTARADAPPYGIDYPTHRPTGRFSNGLNIPDLISLELGLKPTLP-YLSPLL 183
Query: 100 -GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYL 158
G+ +L G NFASA GI +TG Q I Q++ + ++ +G E N
Sbjct: 184 VGEKLLIGANFASAGIGILNDTGIQFLNIIHIQKQLKLFHEYQERLSLHIGAEG-TRNLX 360
Query: 159 SKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVL 218
++ + I LG ND++NNY++ + + SRQ++ Y LI Y + L+ LY+ GAR++++
Sbjct: 361 NRALVLITLGGNDFVNNYYLVPYSARSRQFSLPDYVRYLISEYRKVLRRLYDLGARRVLV 540
Query: 219 FGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAY 278
G G +GC P ELA RS+ G C ++ A +FN +L +++ N +L I NA
Sbjct: 541 TGTGPMGCVPAELATRSRTG-DCDVELQRAASLFNPQLVQMLNGLNQELGADVFIAANAQ 717
Query: 279 GIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGN 338
+ D +++P YGF + CCG G NG C P C NR Y FWD FHPSE +
Sbjct: 718 RMHMDFVSNPRAYGFVTSKIACCGQGPYNGVGLCTPTSNLCPNRDLYAFWDPFHPSEKAS 897
Query: 339 VVIAQR 344
+I Q+
Sbjct: 898 RIIVQQ 915
>TC207231 similar to UP|Q94CH6 (Q94CH6) Family II lipase EXL3, partial (70%)
Length = 1216
Score = 199 bits (506), Expect = 2e-51
Identities = 102/302 (33%), Positives = 171/302 (55%), Gaps = 3/302 (0%)
Frame = +3
Query: 37 GDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPY 95
GDS+VD GNNN +++L + D+ PYG DF GG P+GRF NGK D++ E LG + +P Y
Sbjct: 3 GDSIVDPGNNNKVKTLVKCDFPPYGKDFEGGIPTGRFCNGKIPSDLLVEELGIKELLPAY 182
Query: 96 VSTS--GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQ 153
+ + D++ GV FAS A+G T ++ IS S Q+ ++ + ++ +I+G ED+
Sbjct: 183 LDPNLKPSDLVTGVCFASGASGYDPLT-PKIASVISMSEQLDMFKEYIGKLKHIVG-EDR 356
Query: 154 AANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGA 213
L+ + + GS+D N YF+ + QY Y D+++ + + ++ LY GA
Sbjct: 357 TKFILANSFFLVVAGSDDIANTYFIARVRQL--QYDIPAYTDLMLHSASNFVKELYGLGA 530
Query: 214 RKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVI 273
R++ + IGC P++ R C + N A ++FN+KL +D + LP++R++
Sbjct: 531 RRIGVLSAPPIGCVPSQRTLAGGFQRECAEEYNYAAKLFNSKLSRELDALKHNLPNSRIV 710
Query: 274 YINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHP 333
YI+ Y DII + +G+ + GCCG G+ + C P+ C + +Y+FWD++HP
Sbjct: 711 YIDVYNPLMDIIVNYQRHGYKVVDRGCCGTGKLEVAVLCNPLGATCPDASQYVFWDSYHP 890
Query: 334 SE 335
+E
Sbjct: 891 TE 896
>TC205733
Length = 1377
Score = 197 bits (501), Expect = 6e-51
Identities = 116/335 (34%), Positives = 179/335 (52%), Gaps = 5/335 (1%)
Frame = +2
Query: 14 VMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS-GRF 72
++VL L + +V +FGDS VD GNNN + ++AR+++ PYG DF GG + GRF
Sbjct: 104 IVVLLLLSLVAETSAKVSAVIVFGDSSVDAGNNNFIPTIARSNFQPYGRDFEGGKATGRF 283
Query: 73 SNGKTTVDVIAELLGFDDFIPPYVSTSGD--DILRGVNFASAAAGIREETGQQLGGRISF 130
NG+ D I+E G ++P Y+ + D GV FASAA G T L I
Sbjct: 284 CNGRIPTDFISESFGLKPYVPAYLDPKYNISDFASGVTFASAATGYDNATSDVLSV-IPL 460
Query: 131 SGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYF-MPQFYSTSRQYT 189
Q++ Y+ + LG E +A + +++ ++ + LG+ND+L NY+ MP + Q+T
Sbjct: 461 WKQLEYYKGYQKNLSAYLG-ESKAKDTIAEALHLMSLGTNDFLENYYTMP---GRASQFT 628
Query: 190 TDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDAN 249
QY + L +++LY GARK+ L G+ +GC P E G CV+ N+
Sbjct: 629 PQQYQNFLAGIAENFIRSLYGLGARKVSLGGLPPMGCLPLERTTSIAGGNDCVARYNNIA 808
Query: 250 QIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQ 309
FNN+LK++ + N +LP ++++ N Y I II P YGF +T+ CC G
Sbjct: 809 LEFNNRLKNLTIKLNQELPGLKLVFSNPYYIMLSIIKRPQLYGFESTSVACCATGMFEMG 988
Query: 310 ITCLPMQT-PCENRREYLFWDAFHPSEAGNVVIAQ 343
C Q C + +Y+FWD+FHP+E N ++A+
Sbjct: 989 YACSRGQMFSCTDASKYVFWDSFHPTEMTNSIVAK 1093
>CA800242
Length = 409
Score = 179 bits (453), Expect = 2e-45
Identities = 85/134 (63%), Positives = 106/134 (78%)
Frame = +2
Query: 102 DILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKC 161
DI GVN+ASAA+GIR+ETGQQLG RIS GQVQN+ T Q++N LG+ ++ YL +C
Sbjct: 5 DIFYGVNYASAASGIRDETGQQLGSRISLRGQVQNHIRTAYQMLNSLGDVNRTLTYLGRC 184
Query: 162 IYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGI 221
IYSIG+G +DYLNNYFMPQFY T QYT +QYA++L+Q+Y + L+ LYN+GARKMVLFGI
Sbjct: 185 IYSIGVGGDDYLNNYFMPQFYPTFLQYTPEQYANLLLQSYAQLLEVLYNYGARKMVLFGI 364
Query: 222 GQIGCSPNELAQRS 235
IGC+P LAQ S
Sbjct: 365 SPIGCTPYALAQSS 406
>AW348454 similar to GP|21537293|gb GDSL-motif lipase/hydrolase-like protein
{Arabidopsis thaliana}, partial (63%)
Length = 763
Score = 175 bits (443), Expect = 3e-44
Identities = 87/226 (38%), Positives = 145/226 (63%), Gaps = 4/226 (1%)
Frame = -3
Query: 133 QVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQ 192
Q+ N++ T +V++ E A + ++ Y IG+GSNDY+NN+ P F + +QYT D+
Sbjct: 755 QINNFKKT-KEVISXNIXEAAANKHCNEATYFIGIGSNDYVNNFLQP-FLADGQQYTHDE 582
Query: 193 YADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIF 252
+ ++LI +QLQ+LY GARK+V G+G +GC P++ +S+ G+ C+ +N+ F
Sbjct: 581 FIELLISTLDQQLQSLYQLGARKIVFHGLGPLGCIPSQRV-KSKRGQ-CLKRVNEWILQF 408
Query: 253 NNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITC 312
N+ ++ +++ N++LP+A+ I+ + Y + D+I +P+TYGF +N CC V + G + C
Sbjct: 407 NSNVQKLINTLNHRLPNAKFIFADTYPLVLDLINNPSTYGFKVSNTSCCNVDTSIGGL-C 231
Query: 313 LPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAY----SAQSPSDA 354
LP C NR E++FWDAFHPS+A N V+A++ + SA SP+ A
Sbjct: 230 LPNSKVCRNRHEFVFWDAFHPSDAANAVLAEKFFSLFSSAPSPASA 93
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,779,900
Number of Sequences: 63676
Number of extensions: 189452
Number of successful extensions: 1204
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 1047
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1053
length of query: 365
length of database: 12,639,632
effective HSP length: 99
effective length of query: 266
effective length of database: 6,335,708
effective search space: 1685298328
effective search space used: 1685298328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0404.11


