

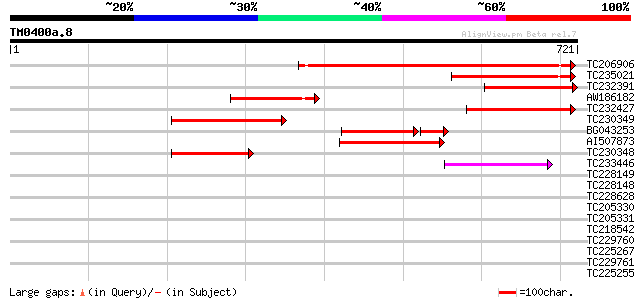
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.8
(721 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206906 similar to UP|Q6Z901 (Q6Z901) Kinesin light chain-like ... 466 e-131
TC235021 similar to UP|Q7XA91 (Q7XA91) At3g27960, partial (23%) 221 1e-57
TC232391 similar to UP|Q7XA91 (Q7XA91) At3g27960, partial (14%) 210 2e-54
AW186182 similar to GP|9294312|dbj contains similarity to kinesi... 207 1e-53
TC232427 similar to UP|Q6Z901 (Q6Z901) Kinesin light chain-like ... 201 1e-51
TC230349 weakly similar to UP|Q15287 (Q15287) RNA-binding protei... 197 1e-50
BG043253 141 2e-44
AI507873 164 1e-40
TC230348 140 2e-33
TC233446 similar to UP|Q6K715 (Q6K715) Kinesin light chain-like,... 56 6e-08
TC228149 similar to UP|O02402 (O02402) Insoluble protein, partia... 37 0.024
TC228148 similar to UP|Q7YXC8 (Q7YXC8) C. elegans GRD-1 protein ... 35 0.12
TC228628 weakly similar to UP|Q6Z4S6 (Q6Z4S6) Tetratricopeptide ... 33 0.34
TC205330 similar to GB|BAA98195.1|8978342|AP002030 short chain a... 33 0.44
TC205331 similar to GB|BAA98195.1|8978342|AP002030 short chain a... 33 0.44
TC218542 31 1.7
TC229760 similar to UP|Q9C566 (Q9C566) Cyclophilin-40 (Expressed... 31 1.7
TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucos... 31 1.7
TC229761 similar to UP|Q9C566 (Q9C566) Cyclophilin-40 (Expressed... 31 1.7
TC225255 similar to UP|Q7XXS4 (Q7XXS4) Thiamine biosynthetic enz... 31 2.2
>TC206906 similar to UP|Q6Z901 (Q6Z901) Kinesin light chain-like protein,
partial (58%)
Length = 1342
Score = 466 bits (1200), Expect = e-131
Identities = 238/352 (67%), Positives = 290/352 (81%)
Frame = +1
Query: 368 LEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASV 427
LEIHR + AS+EE+ADRRLM LI ++KGDYE+ALEH VLASMAM ANGQ+ +VAS+
Sbjct: 25 LEIHRAHSE--PASLEEAADRRLMALICEAKGDYESALEHLVLASMAMIANGQDNEVASI 198
Query: 428 DSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSK 487
D SIG+ Y++L R+DEA+FSYQKALTVFKS KGENHP+VASV+VRLADLY++ GK ++SK
Sbjct: 199 DVSIGNIYMSLCRFDEAIFSYQKALTVFKSAKGENHPSVASVFVRLADLYHRTGKLRESK 378
Query: 488 SYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQ 547
SYCENALRI+ K PG EEIA GL +++A+++S+++ E+ LKLL +A+K+ PGQQ
Sbjct: 379 SYCENALRIYSKPVPGTTAEEIAGGLTEISAVFESVDEPEEALKLLNRAMKLLEDKPGQQ 558
Query: 548 STIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRY 607
STIAGIEA+MGVMYYM+G Y DS N F+SAVAK RASGE+KSA FG+ LNQMGLACVQ +
Sbjct: 559 STIAGIEARMGVMYYMIGRYEDSRNSFESAVAKLRASGERKSAFFGVVLNQMGLACVQLF 738
Query: 608 AINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREE 667
I+EAA+LFEEAR ILE+E G H DTLGVYSNLA TYDAMGRV DAIEILE+V+ +REE
Sbjct: 739 KIDEAAELFEEARGILEQECGPCHQDTLGVYSNLAATYDAMGRVGDAIEILEYVLKLREE 918
Query: 668 KLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIK 719
KLG ANPD +DEKRRLAELLKEAG+ R+RK+K SLE L+D S+ K K
Sbjct: 919 KLGIANPDFEDEKRRLAELLKEAGKTRDRKAK-SLENLIDPGSKRTKKEGAK 1071
>TC235021 similar to UP|Q7XA91 (Q7XA91) At3g27960, partial (23%)
Length = 686
Score = 221 bits (562), Expect = 1e-57
Identities = 112/157 (71%), Positives = 131/157 (83%)
Frame = +1
Query: 563 MLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTI 622
MLGNYS SYN K+A++K RA GEKKS+ FGIALNQMGLACVQ YA++EA +LFEEA++
Sbjct: 1 MLGNYSKSYNTLKNAISKLRAIGEKKSSFFGIALNQMGLACVQCYALSEATELFEEAKSF 180
Query: 623 LEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRR 682
LE EYG YHP+TLGV SNLA TYDA+GR+DDAI+ILE+VV REEKLGTANP+VDDEKRR
Sbjct: 181 LEHEYGPYHPETLGVSSNLAATYDAIGRLDDAIQILEYVVNTREEKLGTANPEVDDEKRR 360
Query: 683 LAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIK 719
L ELLKEAGR R+RK+ RSLE LLD N+ N I+
Sbjct: 361 LGELLKEAGRVRSRKT-RSLENLLDGNAHAANNVVIR 468
>TC232391 similar to UP|Q7XA91 (Q7XA91) At3g27960, partial (14%)
Length = 562
Score = 210 bits (535), Expect = 2e-54
Identities = 105/118 (88%), Positives = 112/118 (93%)
Frame = +3
Query: 604 VQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVG 663
VQ YAINEAADLFEEARTILEKEYG YHPDTLGVYSNLAGTYDAMGRVDDAIEILE+VVG
Sbjct: 3 VQCYAINEAADLFEEARTILEKEYGPYHPDTLGVYSNLAGTYDAMGRVDDAIEILEYVVG 182
Query: 664 MREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIKVL 721
MREEKLGTANPDVDDEKRRL ELLKE+GR RNR+S+RSLETLLD +S+L+KNN IKVL
Sbjct: 183 MREEKLGTANPDVDDEKRRLEELLKESGRARNRRSRRSLETLLDTHSQLVKNNGIKVL 356
>AW186182 similar to GP|9294312|dbj contains similarity to kinesin light
chain~gene_id:K24A2.5 {Arabidopsis thaliana}, partial
(16%)
Length = 333
Score = 207 bits (527), Expect = 1e-53
Identities = 102/112 (91%), Positives = 108/112 (96%)
Frame = +3
Query: 282 SIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFG 341
SIDIPVLEDGQDHALAKFAGCMQLGDTYAMMG IENS+LFYTAGLEIQGQVLGETDPRFG
Sbjct: 3 SIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGQIENSLLFYTAGLEIQGQVLGETDPRFG 182
Query: 342 ETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGL 393
ETCRYVAEAHVQALQFDEAE+LCQMAL+IHR GNG+ AS+EE+ADRRLMGL
Sbjct: 183 ETCRYVAEAHVQALQFDEAEKLCQMALDIHR--GNGAPASIEEAADRRLMGL 332
>TC232427 similar to UP|Q6Z901 (Q6Z901) Kinesin light chain-like protein,
partial (24%)
Length = 504
Score = 201 bits (510), Expect = 1e-51
Identities = 101/138 (73%), Positives = 115/138 (83%)
Frame = +2
Query: 582 RASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNL 641
RASGEKKSA FG+ LNQMGLACVQ Y I +AA FEEA+ ILE+E G YH DTLGVYSNL
Sbjct: 5 RASGEKKSAFFGVVLNQMGLACVQLYKIGDAAKHFEEAKEILERECGTYHSDTLGVYSNL 184
Query: 642 AGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRS 701
A TYDA+GRV+DAIEILE+++ MREEKLGTANPDVDDEK+RL ELLKEAGR RNRK K+S
Sbjct: 185 AATYDALGRVEDAIEILEYILKMREEKLGTANPDVDDEKKRLFELLKEAGRVRNRKGKKS 364
Query: 702 LETLLDANSRLIKNNSIK 719
LE L+D+NS +K K
Sbjct: 365 LENLIDSNSLKMKKEGKK 418
>TC230349 weakly similar to UP|Q15287 (Q15287) RNA-binding protein S1,
serine-rich domain, partial (12%)
Length = 479
Score = 197 bits (502), Expect = 1e-50
Identities = 94/148 (63%), Positives = 120/148 (80%), Gaps = 1/148 (0%)
Frame = +1
Query: 206 LDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCA-DGKPSLELVMCLHVLAT 264
LDNPDLGPFLLK RD I+SG+ P KALD A+RA KSFE CA +G+PSL+L M LHVLA
Sbjct: 34 LDNPDLGPFLLKLARDTIASGDGPSKALDFAIRASKSFERCAVEGEPSLDLAMSLHVLAA 213
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
IYC+LG+++EA+P+LER+I +P +E G DHALA F+G MQLGDT++M+G ++ SI Y
Sbjct: 214 IYCSLGRFDEAVPVLERAIQVPDVERGADHALAAFSGYMQLGDTFSMLGQVDRSISCYDQ 393
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEAHV 352
GL+IQ Q LG++DPR GETCRY+ EA+V
Sbjct: 394 GLQIQIQALGDSDPRVGETCRYLTEANV 477
Score = 35.4 bits (80), Expect = 0.089
Identities = 24/75 (32%), Positives = 40/75 (53%), Gaps = 5/75 (6%)
Frame = +1
Query: 435 YLALARYDEAVFSYQKALTVFKSTKGENHPNVA-SVYVRLADLYNKIGKFKDSKSYCENA 493
Y +L R+DEAV ++A+ V +G +H A S Y++L D ++ +G+ S S +
Sbjct: 217 YCSLGRFDEAVPVLERAIQVPDVERGADHALAAFSGYMQLGDTFSMLGQVDRSISCYDQG 396
Query: 494 LRI----FGKSKPGI 504
L+I G S P +
Sbjct: 397 LQIQIQALGDSDPRV 441
>BG043253
Length = 411
Score = 141 bits (356), Expect(2) = 2e-44
Identities = 66/97 (68%), Positives = 83/97 (85%)
Frame = +2
Query: 423 DVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGK 482
+VASVD SIGD YL+L+RYDEAVF+YQKALTVFK++KGENHP V V++RLADLYN+ GK
Sbjct: 5 EVASVDCSIGDTYLSLSRYDEAVFAYQKALTVFKTSKGENHPAVGLVFLRLADLYNRTGK 184
Query: 483 FKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAI 519
++SKSYCE+AL+I+ PGIPPEEIA+GL ++ I
Sbjct: 185 IRESKSYCESALKIYENPMPGIPPEEIASGLTNILTI 295
Score = 57.0 bits (136), Expect(2) = 2e-44
Identities = 28/35 (80%), Positives = 30/35 (85%)
Frame = +3
Query: 523 MNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQM 557
MNDLE LKLL+KAL+IY PGQQSTIAGIEAQM
Sbjct: 306 MNDLEHALKLLQKALEIYNDTPGQQSTIAGIEAQM 410
>AI507873
Length = 405
Score = 164 bits (416), Expect = 1e-40
Identities = 79/133 (59%), Positives = 106/133 (79%)
Frame = +2
Query: 420 QEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNK 479
++ +VA++D SIGD YL+L R+DEAVF+YQKALTVFKSTKGE+H VA VY+RLADLY +
Sbjct: 5 EDNEVAAIDVSIGDIYLSLCRFDEAVFAYQKALTVFKSTKGESHSCVALVYIRLADLYYR 184
Query: 480 IGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKI 539
GK ++SKSYCENALRI+ K G EIA+GL +++AIY+++N+ E+ LKLL+KA+K+
Sbjct: 185 TGKLRESKSYCENALRIYSKPVAGTTAGEIASGLTEISAIYEALNEPEEALKLLQKAVKL 364
Query: 540 YGHAPGQQSTIAG 552
PGQ T+AG
Sbjct: 365 LEDIPGQYRTVAG 403
>TC230348
Length = 438
Score = 140 bits (354), Expect = 2e-33
Identities = 68/105 (64%), Positives = 84/105 (79%), Gaps = 1/105 (0%)
Frame = +1
Query: 206 LDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCA-DGKPSLELVMCLHVLAT 264
LDNPDLGPFLLK RD I+SG+ P KALD A+RA SFE CA G+PSL+L M LHVLA
Sbjct: 124 LDNPDLGPFLLKLARDTIASGDGPAKALDFAIRASTSFERCAIQGEPSLDLAMSLHVLAA 303
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTY 309
IYC+LG++ EA+P+LER+I +P ++ G DHALA F+G MQLGDT+
Sbjct: 304 IYCSLGRFEEAVPVLERAILVPDVDRGPDHALASFSGYMQLGDTF 438
>TC233446 similar to UP|Q6K715 (Q6K715) Kinesin light chain-like, partial
(22%)
Length = 698
Score = 55.8 bits (133), Expect = 6e-08
Identities = 37/138 (26%), Positives = 64/138 (45%)
Frame = +1
Query: 553 IEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEA 612
+ A++G + + G + +SA + + S K G N +G A ++ A
Sbjct: 13 VSARIGWLLLLTGKVQQAIPYLESAAERLKDSFGPKHFGVGYIYNNLGAAYLELDRPQSA 192
Query: 613 ADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTA 672
A +F A+ I++ G +H DT+ NL+ Y MG AIE + VV E +A
Sbjct: 193 AQMFAVAKDIMDTSLGPHHADTIEACQNLSKAYGEMGSYVLAIEFQQQVVDAWESHGASA 372
Query: 673 NPDVDDEKRRLAELLKEA 690
++ + +R L +L K+A
Sbjct: 373 EDELREGQRLLDQLKKKA 426
>TC228149 similar to UP|O02402 (O02402) Insoluble protein, partial (4%)
Length = 1008
Score = 37.4 bits (85), Expect = 0.024
Identities = 30/114 (26%), Positives = 49/114 (42%), Gaps = 1/114 (0%)
Frame = -2
Query: 69 VCEMRSSDQS-PSRASFYSYGEESRIDSELGHLVGGILEITKEVVTENKEESNGNAAEKD 127
VC RSS S PS A+F G++ + + K ++ EE A EK
Sbjct: 983 VCRKRSSMSSKPSSATFGQKGDQPVFNDKKERPSSDAKPPMKPSSSDTDEEELPKAKEKP 804
Query: 128 IVSCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKG 181
+ N S+S ++ + S+ STPK+ + +PS + E+S +G
Sbjct: 803 VTGARSLRRSNKNLNSNSSNNNNKKPSSISTPKAGSSWNKPSETVKPEKSKAEG 642
>TC228148 similar to UP|Q7YXC8 (Q7YXC8) C. elegans GRD-1 protein
(Corresponding sequence R08B4.1b), partial (3%)
Length = 1180
Score = 35.0 bits (79), Expect = 0.12
Identities = 34/129 (26%), Positives = 54/129 (41%), Gaps = 4/129 (3%)
Frame = +3
Query: 69 VCEMRSSDQS-PSRASFYSYGEESRIDSELGHLVGGILEITKEVVTENKEESNGNAAEKD 127
VC RSS + PS A+F G++ + + K ++ EE A EK
Sbjct: 81 VCRKRSSMLAKPSSATFGQKGDQPTFNDKKERPSSDAKPPMKPSSSDTDEEELPKAKEKP 260
Query: 128 IV---SCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLY 184
+ S + +NN + PSS STPK+ + +PS + E+S +G
Sbjct: 261 VTGARSLRRSYKNLNNNNNKKPSSN-------STPKTGSSGNKPSETVKPEKSKAEGG-- 413
Query: 185 PMRKHRSLA 193
P +K + A
Sbjct: 414 PDKKRNAAA 440
>TC228628 weakly similar to UP|Q6Z4S6 (Q6Z4S6) Tetratricopeptide
repeat-containing protein-like, partial (25%)
Length = 920
Score = 33.5 bits (75), Expect = 0.34
Identities = 34/180 (18%), Positives = 69/180 (37%), Gaps = 23/180 (12%)
Frame = +2
Query: 429 SSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKS 488
S++ D L ++ ++ YQKALT+ + + +A + R+ K +++ +
Sbjct: 32 STLADVALEREDFETSLSDYQKALTILEQLVEPDDRKIADLNFRICLCLEVSSKPQEAIA 211
Query: 489 YCENALRI-----------------------FGKSKPGIPPEEIANGLIDVAAIYQSMND 525
YC+ A + + P P E N ++D + +++
Sbjct: 212 YCQKATSVCKARLHRLTNEVKSCSDLTSASELAQDVPACPKSESNNSILDKQSEIETLKG 391
Query: 526 LEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASG 585
L L+ KK + +S +A I +G+ GN +S + S+ A+G
Sbjct: 392 LSSELE--KKLEDLQQLVSNPKSILAEI---LGIAAAKAGNVKESSSAMVSSSQLATANG 556
>TC205330 similar to GB|BAA98195.1|8978342|AP002030 short chain alcohol
dehydrogenase-like {Arabidopsis thaliana;} , partial
(70%)
Length = 661
Score = 33.1 bits (74), Expect = 0.44
Identities = 17/52 (32%), Positives = 26/52 (49%), Gaps = 2/52 (3%)
Frame = +3
Query: 343 TCRYVAEAH--VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMG 392
T Y AE + + A D LCQ+A + + +GNGS S+ A + +G
Sbjct: 348 TIEYTAEEYSKLMATNLDSTYHLCQLAYPLLKASGNGSIVSISSVASQTSVG 503
>TC205331 similar to GB|BAA98195.1|8978342|AP002030 short chain alcohol
dehydrogenase-like {Arabidopsis thaliana;} , partial
(85%)
Length = 921
Score = 33.1 bits (74), Expect = 0.44
Identities = 17/52 (32%), Positives = 26/52 (49%), Gaps = 2/52 (3%)
Frame = +2
Query: 343 TCRYVAEAH--VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMG 392
T Y AE + + A D LCQ+A + + +GNGS S+ A + +G
Sbjct: 320 TIEYTAEEYSKLMATNLDSTYHLCQLAYPLLKASGNGSIVSISSVASQTSVG 475
>TC218542
Length = 1167
Score = 31.2 bits (69), Expect = 1.7
Identities = 31/91 (34%), Positives = 41/91 (44%), Gaps = 7/91 (7%)
Frame = +1
Query: 560 MYYMLGNYSDSY--NIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAAD--- 614
MYY +G+ S+S+ NIF S + G S AL++M C+QRY +D
Sbjct: 28 MYY-IGSLSESHLQNIFFSYGMAYGGGGFAISYPLAKALSKMQDRCIQRYPALYGSDDRM 204
Query: 615 --LFEEARTILEKEYGQYHPDTLGVYSNLAG 643
E L KE G + D VY NL G
Sbjct: 205 QACMAELGVPLTKEIGFHQYD---VYGNLFG 288
>TC229760 similar to UP|Q9C566 (Q9C566) Cyclophilin-40 (Expressed protein),
partial (70%)
Length = 1164
Score = 31.2 bits (69), Expect = 1.7
Identities = 21/54 (38%), Positives = 27/54 (49%)
Frame = +2
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKD 485
G AY+AL D AV S++KALT+ PN A + LA KI +D
Sbjct: 620 GQAYMALHDIDAAVESFKKALTL--------EPNDAGIKKELAAARKKIADRRD 757
>TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucosidase,
partial (69%)
Length = 2006
Score = 31.2 bits (69), Expect = 1.7
Identities = 24/58 (41%), Positives = 29/58 (49%), Gaps = 3/58 (5%)
Frame = +3
Query: 200 ERIATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEM---CADGKPSLE 254
E I GLD D GPFL T I G + L+LAL L S +M DG+PS +
Sbjct: 366 EAIKAGLDL-DCGPFLAIHTDSAIRKGLISENDLNLALANLISVQMRLGMFDGEPSTQ 536
>TC229761 similar to UP|Q9C566 (Q9C566) Cyclophilin-40 (Expressed protein),
partial (23%)
Length = 523
Score = 31.2 bits (69), Expect = 1.7
Identities = 21/54 (38%), Positives = 27/54 (49%)
Frame = +1
Query: 432 GDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKD 485
G AY+AL D AV S++KALT+ PN A + LA KI +D
Sbjct: 82 GQAYMALHDIDAAVESFKKALTL--------EPNDAGIKKELAAARKKIADRRD 219
>TC225255 similar to UP|Q7XXS4 (Q7XXS4) Thiamine biosynthetic enzyme, partial
(87%)
Length = 1299
Score = 30.8 bits (68), Expect = 2.2
Identities = 18/63 (28%), Positives = 29/63 (45%), Gaps = 4/63 (6%)
Frame = -1
Query: 139 DNNQSHS----PSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLAL 194
DN+ H+ P + ++ + SRN P+T + S KGS+ P R H +L L
Sbjct: 906 DNHARHNLPSEPDNSVLSSIHIQSLHSRNAINHPNTLQPLNPSGAKGSIVPTRAHHNLRL 727
Query: 195 RGI 197
+
Sbjct: 726 HHV 718
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,007,673
Number of Sequences: 63676
Number of extensions: 274223
Number of successful extensions: 1047
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 1035
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1042
length of query: 721
length of database: 12,639,632
effective HSP length: 104
effective length of query: 617
effective length of database: 6,017,328
effective search space: 3712691376
effective search space used: 3712691376
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0400a.8


