

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
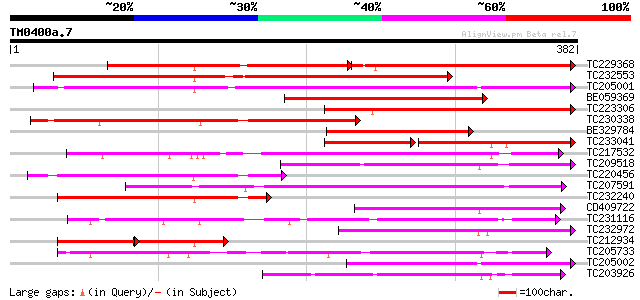
Query= TM0400a.7
(382 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229368 similar to PIR|T48618|T48618 early nodule-specific prot... 169 6e-86
TC232553 similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (59%) 249 2e-66
TC205001 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetyles... 248 3e-66
BE059369 weakly similar to PIR|T09416|T094 coil protein PO22 mi... 222 2e-58
TC223306 weakly similar to PIR|T48618|T48618 early nodule-specif... 197 5e-51
TC230338 similar to UP|Q8W0Y6 (Q8W0Y6) Enod8.2, partial (49%) 182 2e-46
BE329784 161 5e-40
TC233041 weakly similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment)... 109 2e-37
TC217532 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 147 8e-36
TC209518 weakly similar to UP|O82076 (O82076) Lipase homolog (Fr... 141 6e-34
TC220456 weakly similar to UP|O81262 (O81262) Early nodule-speci... 140 1e-33
TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 125 2e-29
TC232240 weakly similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial ... 125 4e-29
CD409722 124 7e-29
TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 112 2e-25
TC232972 weakly similar to UP|O82681 (O82681) Lanatoside 15'-O-a... 110 1e-24
TC212934 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetyles... 72 3e-24
TC205733 98 7e-21
TC205002 similar to UP|Q9XIY6 (Q9XIY6) ESTs AU075322(C11109), pa... 97 1e-20
TC203926 weakly similar to PIR|T06696|T06696 lipase homolog T29H... 97 1e-20
>TC229368 similar to PIR|T48618|T48618 early nodule-specific protein-like -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(61%)
Length = 1238
Score = 169 bits (429), Expect(2) = 6e-86
Identities = 84/155 (54%), Positives = 106/155 (68%), Gaps = 4/155 (2%)
Frame = +3
Query: 231 PIGCLPTSSIFYEPK--KGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYV 288
P GC+P +FY+ +G LD GCV N +A EFN+QLKD+V +LR LP+A TYV
Sbjct: 504 PFGCMPVQ-LFYKHNIPEGYLDQYGCVKDQNVMATEFNKQLKDRVIKLRTELPEAAITYV 680
Query: 289 DVYTAKYELISNASKQGFVNPLEVCCGSYYG-YRIDCGKKAVVNG-TVYGNPCKNPSQHI 346
DVY AKY LISN K+GFV+P+++CCG + I CG NG V+G+ C+NPSQ+I
Sbjct: 681 DVYAAKYALISNTKKEGFVDPMKICCGYHVNDTHIWCGNLGTDNGKDVFGSACENPSQYI 860
Query: 347 SWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
SWD VHY +AAN WVA I +GS +DPP PI QAC
Sbjct: 861 SWDSVHYAEAANHWVANRILNGSYTDPPTPITQAC 965
Score = 166 bits (420), Expect(2) = 6e-86
Identities = 85/171 (49%), Positives = 110/171 (63%), Gaps = 6/171 (3%)
Frame = +2
Query: 67 GISFFGNLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRP--- 123
G FF SGR DGRLI+DFI E+L +PYLSAYLNS+G+NYRHGANFA GG++IR
Sbjct: 5 GEGFFHKPSGRDCDGRLIVDFIAEKLNLPYLSAYLNSLGTNYRHGANFATGGSTIRKQNE 184
Query: 124 ---VYGFSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTID 180
YG SPF L +Q+ QF Q ++ + L + + P KS LP PE+FSKALYT D
Sbjct: 185 TIFQYGISPFSLDIQIVQFNQFKARTKQLYEEAKA----PHEKSKLPVPEEFSKALYTFD 352
Query: 181 IGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGP 231
IGQNDL G + +++ S+P+++ V+ +Y G R FWIHNT P
Sbjct: 353 IGQNDLSVGFRKMNFDQIRESMPDILNQLANAVKNIYQQGGRYFWIHNTSP 505
>TC232553 similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (59%)
Length = 832
Score = 249 bits (635), Expect = 2e-66
Identities = 129/277 (46%), Positives = 178/277 (63%), Gaps = 8/277 (2%)
Frame = +2
Query: 30 SYSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFIT 89
S ++S +C +PA++NFGDSNSDTG + AAF P G S+F + +GR DGRLI+DF+
Sbjct: 8 SLAASKQCHFPAIFNFGDSNSDTGGLSAAFGQAGPPHGESYFHHPAGRYCDGRLIVDFLA 187
Query: 90 EELEIPYLSAYLNSIGSNYRHGANFAAGGASIRP-------VYGFSPFYLGMQVAQFIQL 142
++L +PYLSA+L+S+GSNY HGANFA G++IRP GFSPF L +Q QF
Sbjct: 188 KKLGLPYLSAFLDSVGSNYSHGANFATAGSTIRPQNTTLHQTGGFSPFSLDVQFNQFSDF 367
Query: 143 QSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHT-SEEEVLRS 201
Q QF N+ +K+ LP+ EDFS+ALYT DIGQNDL G H S ++V
Sbjct: 368 QRR-----TQFFHNK-GGVYKTLLPKAEDFSQALYTFDIGQNDLASGYFHNMSTDQVKAY 529
Query: 202 IPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKI 261
+P+++ F ++ +Y+ G R FW+HNTGP+GCLP + K +D C P+N++
Sbjct: 530 VPDVLAQFKNVIKYVYNHGGRSFWVHNTGPVGCLPYIMDLHPVKPSLVDKAXCATPYNEV 709
Query: 262 AQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELI 298
A+ FN +LK+ V QL + LP A TYVDVY+ K+ LI
Sbjct: 710 AKFFNSKLKEVVVQLTKELPLAAITYVDVYSVKHSLI 820
>TC205001 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetylesterase
precursor, partial (62%)
Length = 1384
Score = 248 bits (633), Expect = 3e-66
Identities = 136/372 (36%), Positives = 203/372 (54%), Gaps = 7/372 (1%)
Frame = +1
Query: 17 VACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSG 76
V C + I + SYS C + A++NFGDSNSDTG + +F +P G+++F G
Sbjct: 100 VICMVMMISLVDSSYSL---CDFEAIFNFGDSNSDTGGFHTSFPAQPAPYGMTYFKKPVG 270
Query: 77 RASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPF 130
RASDGRLI+DF+ + L +PYLS YL SIGS+Y HGANFA+ +++ P V G SPF
Sbjct: 271 RASDGRLIVDFLAQGLGLPYLSPYLQSIGSDYTHGANFASSASTVIPPTTSFSVSGLSPF 450
Query: 131 YLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGL 190
L +Q+ Q Q ++ ++ +F T + +P P+ F KALYT IGQND +
Sbjct: 451 SLSVQLRQMEQFKAKVD----EFHQTGTRISSGTKIPSPDIFGKALYTFYIGQNDFTSKI 618
Query: 191 MHTSEEEVLR-SIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNL 249
T + +R S+P ++ ++ LY G R F + N GP+GC P + +
Sbjct: 619 AATGGIDGVRGSLPHIVSQINAAIKELYAQGGRAFMVFNLGPVGCYPGYLVELPHATSDY 798
Query: 250 DANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNP 309
D GC++ HN ++N+ L+D + Q +L A Y D ++A EL + + G
Sbjct: 799 DEFGCIVSHNNAVNDYNKLLRDTLTQTGESLVDASLIYADTHSALLELFHHPTFYGLKYN 978
Query: 310 LEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGS 369
CCG YG + ++ G + + C P ++SWDG+H+T+AANK VA I +GS
Sbjct: 979 TRTCCG--YGGGVYNFNPKILCGHMLASACDEPQNYVSWDGIHFTEAANKIVAHAILNGS 1152
Query: 370 LSDPPVPIGQAC 381
L PP P+ + C
Sbjct: 1153LFYPPFPLHKHC 1188
>BE059369 weakly similar to PIR|T09416|T094 coil protein PO22
microspore/pollen-specific - alfalfa, partial (15%)
Length = 411
Score = 222 bits (565), Expect = 2e-58
Identities = 103/137 (75%), Positives = 115/137 (83%)
Frame = +1
Query: 186 LGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPK 245
L FGL HTS+E+V++SIPE++ F VQ LY+V ARVFWIHNTGPIGCLP S I+YEPK
Sbjct: 1 LAFGLQHTSQEQVIKSIPEILNQFFQAVQQLYNVRARVFWIHNTGPIGCLPYSYIYYEPK 180
Query: 246 KGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQG 305
KGN+DANGCV P N +AQEFNRQLKDQVFQLRR P AKFTYVDVYTAKYELI+N QG
Sbjct: 181 KGNIDANGCVKPQNDLAQEFNRQLKDQVFQLRRKFPLAKFTYVDVYTAKYELINNTRNQG 360
Query: 306 FVNPLEVCCGSYYGYRI 322
FV+PLE CCGSYYGY I
Sbjct: 361 FVSPLEFCCGSYYGYHI 411
>TC223306 weakly similar to PIR|T48618|T48618 early nodule-specific
protein-like - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (25%)
Length = 669
Score = 197 bits (502), Expect = 5e-51
Identities = 92/176 (52%), Positives = 124/176 (70%), Gaps = 7/176 (3%)
Frame = +1
Query: 213 VQVLYDVGARVFWIHNTGPIGCLPTSSIFYE-----PKKGNLDANGCVIPHNKIAQEFNR 267
VQ L +GAR FWIHNTGPIGCLP + + P G LD NGC+ N +A+EFN+
Sbjct: 16 VQTLLGLGARTFWIHNTGPIGCLPVAMPVHNAMNTTPGAGYLDQNGCINYQNDMAREFNK 195
Query: 268 QLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYY-GYRIDCGK 326
+LK+ V +LR P A YVD+++AKYELISNA+K+GFV+P +CCG + GY + CG
Sbjct: 196 KLKNTVVKLRVQFPDASLIYVDMFSAKYELISNANKEGFVDPSGICCGYHQDGYHLYCGN 375
Query: 327 KAVVNG-TVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
KA++NG ++ + C +PS++ISWDGVHYT+AAN W+A I +GS SDPP+ I +C
Sbjct: 376 KAIINGKEIFADTCDDPSKYISWDGVHYTEAANHWIANRILNGSFSDPPLSIAHSC 543
>TC230338 similar to UP|Q8W0Y6 (Q8W0Y6) Enod8.2, partial (49%)
Length = 737
Score = 182 bits (462), Expect = 2e-46
Identities = 97/227 (42%), Positives = 147/227 (64%), Gaps = 5/227 (2%)
Frame = +2
Query: 15 LCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAF--AGLQSPGGISFFG 72
LC+A T + P+ + C +PA++NFG SN+DTG + A+F A +SP G ++F
Sbjct: 83 LCIATTILNNPA---MATKQYYCDFPAIFNFGASNADTGGLAASFFVAAPKSPNGETYFH 253
Query: 73 NLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGF--SPF 130
+GR SDGRLIIDF+ + +PYLS YL+S+G+N+ GA+FA G++I P F SPF
Sbjct: 254 RPAGRFSDGRLIIDFLAQSFGLPYLSPYLDSLGTNFSRGASFATAGSTIIPQQSFRSSPF 433
Query: 131 YLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGL 190
LG+Q +QF + + + + Q F + +P+ E F +ALYT DIGQNDL G
Sbjct: 434 SLGVQYSQFQRFKPTTQFIREQGG------VFATLMPKEEYFHEALYTFDIGQNDLTAGF 595
Query: 191 M-HTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLP 236
+ + ++ +IP+++++FT +++ +Y++GAR FWIHNTGPIGCLP
Sbjct: 596 FGNMTLQQFNATIPDIIKSFTSNIKNIYNMGARSFWIHNTGPIGCLP 736
>BE329784
Length = 454
Score = 161 bits (407), Expect = 5e-40
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +1
Query: 214 QVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQV 273
Q LY+VGARVFWIHNTGPIGCLP S I+YEPKKGN+DANGCV P N +AQEFNRQLKDQV
Sbjct: 7 QQLYNVGARVFWIHNTGPIGCLPYSYIYYEPKKGNIDANGCVKPQNDLAQEFNRQLKDQV 186
Query: 274 FQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEV 312
FQLRR P AKFTYVDVYTAKYELI+N QG L+V
Sbjct: 187 FQLRRKFPLAKFTYVDVYTAKYELINNTRNQGGRQVLKV 303
Score = 35.4 bits (80), Expect = 0.043
Identities = 13/15 (86%), Positives = 14/15 (92%)
Frame = +2
Query: 305 GFVNPLEVCCGSYYG 319
GFV+PLE CCGSYYG
Sbjct: 410 GFVSPLEFCCGSYYG 454
>TC233041 weakly similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment), partial
(35%)
Length = 748
Score = 109 bits (273), Expect(2) = 2e-37
Identities = 57/112 (50%), Positives = 69/112 (60%), Gaps = 6/112 (5%)
Frame = +3
Query: 276 LRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRID----CGKKAVVN 331
LR+ LP A TYVDVYT KY LIS+A K GF + CCG Y + CG VN
Sbjct: 255 LRKELPGAAITYVDVYTVKYTLISHAHKYGFEQGVIACCGHGGKYNFNNTERCGATKRVN 434
Query: 332 GT--VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
GT V N CK+ S I WDG+HYT+AANKW+ + I +GS SDPP + +AC
Sbjct: 435 GTEIVIANSCKDLSVRIIWDGIHYTEAANKWIFQQIVNGSFSDPPHSLKRAC 590
Score = 64.3 bits (155), Expect(2) = 2e-37
Identities = 29/61 (47%), Positives = 38/61 (61%)
Frame = +2
Query: 213 VQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQ 272
++ +Y G FWIHNTGP+GCLP Y K +D GC P N++AQ FNR+LK+
Sbjct: 65 IKGVYGEGGXSFWIHNTGPLGCLPYMLDRYPMKPTQMDEFGCAKPFNEVAQYFNRKLKEV 244
Query: 273 V 273
V
Sbjct: 245 V 247
>TC217532 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(54%)
Length = 1372
Score = 147 bits (371), Expect = 8e-36
Identities = 115/363 (31%), Positives = 176/363 (47%), Gaps = 28/363 (7%)
Frame = +1
Query: 39 YPAVYNFGDSNSDTGVVYAAFAG-----LQSPGGISFFGNLSGRASDGRLIIDFITEELE 93
Y ++++FGDS +DTG +Y L P G + F +GR SDGRLI+DF+ E L
Sbjct: 112 YTSLFSFGDSLTDTGNLYFISPRQSPDCLLPPYGQTHFHRPNGRCSDGRLILDFLAESLG 291
Query: 94 IPYLSAYLNSIGS-----NYRHGANFAAGGASI--RPVY---GFSP-----FYLGMQVAQ 138
+PY+ YL N G NFA GA+ R + GF+ F LG+Q+
Sbjct: 292 LPYVKPYLGFKNGAVKRGNIEQGVNFAVAGATALDRGFFEEKGFAVDVTANFSLGVQLDW 471
Query: 139 FIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTI-DIGQNDLGFGLMHTSE-E 196
F +L + +L N SS + + +L+ + +IG ND G+ L T+
Sbjct: 472 FKEL---LPSLCNSSSSCK------------KVIGSSLFIVGEIGGNDYGYPLSETTAFG 606
Query: 197 EVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTS-SIFYEPKKGNLDANGCV 255
+++ IP+++ T ++ L D+GA F + + P+GC P +IF K D GC+
Sbjct: 607 DLVTYIPQVISVITSAIRELIDLGAVTFMVPGSLPLGCNPAYLTIFATIDKEEYDQAGCL 786
Query: 256 IPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFV-NPLEVCC 314
N + N L+ ++ +LR P Y D + A E ++ + GF N L+VCC
Sbjct: 787 KWLNTFYEYHNELLQIEINRLRVLYPLTNIIYADYFNAALEFYNSPEQFGFGGNVLKVCC 966
Query: 315 GSYYGYRID----CGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSL 370
G Y + CG VV C +PSQ++SWDG H T+AA +W+ K + DG
Sbjct: 967 GGGGPYNYNETAMCGDAGVV-------ACDDPSQYVSWDGYHLTEAAYRWMTKGLLDGPY 1125
Query: 371 SDP 373
+ P
Sbjct: 1126TIP 1134
>TC209518 weakly similar to UP|O82076 (O82076) Lipase homolog (Fragment),
partial (56%)
Length = 874
Score = 141 bits (355), Expect = 6e-34
Identities = 77/205 (37%), Positives = 111/205 (53%), Gaps = 6/205 (2%)
Frame = +1
Query: 183 QNDLGFGLM-HTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIF 241
QNDL + S +V++ IP ++ V+ LY+ GAR FW+HNTGP+GCLP +
Sbjct: 1 QNDLADSFAKNLSYAQVIKKIPAVITEIENAVKNLYNDGARKFWVHNTGPLGCLP--KVL 174
Query: 242 YEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNA 301
+K +LD+ GC+ +N A+ FN L +LR A YVD+Y KY+LI+NA
Sbjct: 175 ALAQKKDLDSLGCLSSYNSAARLFNEALLHSSQKLRSEFKDATLVYVDIYAIKYDLITNA 354
Query: 302 SKQGFVNPLEVCCG-----SYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQA 356
+K GF NPL VCCG + R+ CG+ C ++++SWDG+H T+A
Sbjct: 355 AKYGFSNPLMVCCGYGGPPYNFDVRVTCGQPGY-------QVCDEGARYVSWDGIHQTEA 513
Query: 357 ANKWVAKHIRDGSLSDPPVPIGQAC 381
AN +A I + S P +P C
Sbjct: 514 ANTLIASKILSMAYSTPRIPFDFFC 588
>TC220456 weakly similar to UP|O81262 (O81262) Early nodule-specific protein,
partial (43%)
Length = 587
Score = 140 bits (352), Expect = 1e-33
Identities = 78/180 (43%), Positives = 108/180 (59%), Gaps = 6/180 (3%)
Frame = +1
Query: 13 FALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFG 72
FA+ T + P+ ++ +C +PA++NFGDSNSDTG + A+ P G ++F
Sbjct: 61 FAILSIATIVPNPA-----FATKECVFPAIFNFGDSNSDTGGLAASLIAPTPPYGETYFH 225
Query: 73 NLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIR------PVYG 126
+GR SDGRL+IDFI + +PYLSAYL+S+G+N+ HGANFA ++IR P G
Sbjct: 226 RPAGRFSDGRLVIDFIAKSFGLPYLSAYLDSLGTNFSHGANFATSASTIRLPTSIIPQGG 405
Query: 127 FSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDL 186
FSPFYL +Q QF +S + + +Q F S +P+ E F KALYT DIGQ L
Sbjct: 406 FSPFYLDIQYTQFRDSKSRTQFIRHQGG------VFASLMPKEEYFDKALYTFDIGQKIL 567
>TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(57%)
Length = 1290
Score = 125 bits (315), Expect = 2e-29
Identities = 86/306 (28%), Positives = 145/306 (47%), Gaps = 9/306 (2%)
Frame = +1
Query: 79 SDGRLIIDFITEELEIPYLSAYLN-SIGSNYRHGANFAAGGASIRPVYGFSPFYLGMQVA 137
SDGRL+IDFI E ++PYL YL + + + G NFA GA+ + F++ +A
Sbjct: 4 SDGRLMIDFIAEAYDLPYLPPYLALTKDKDIQRGVNFAVAGATALD----AKFFIEAGLA 171
Query: 138 QFIQLQSHIENLLNQFSSNR-----TEPPFKSYLPRPEDFSKALYTI-DIGQNDLGFGLM 191
+++ + + L F + T+ SY F ++L+ + +IG ND + +
Sbjct: 172 KYLWTNNSLNIQLGWFKKLKPSLCTTKQDCDSY------FKRSLFLVGEIGGNDYNYAAI 333
Query: 192 HTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTS-SIFYEPKKGNLD 250
+ ++ ++P ++ T + L GAR + PIGC ++F K + D
Sbjct: 334 AGNITQLQATVPPVVEAITAAINELIAEGARELLVPGNFPIGCSALYLTLFRSENKEDYD 513
Query: 251 ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN-P 309
+GC+ N A+ N++LK + LR+ P A+ Y D Y A GF N
Sbjct: 514 ESGCLKTFNGFAEYHNKELKLALETLRKKNPHARILYADYYGAAKRFFHAPGHHGFTNGA 693
Query: 310 LEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGS 369
L CCG + + + G+ C +PS + +WDG+H T+AA +++AK + G
Sbjct: 694 LRACCGGGGPFNFNISARCGHTGS---KACADPSTYANWDGIHLTEAAYRYIAKGLIYGP 864
Query: 370 LSDPPV 375
S PP+
Sbjct: 865 FSYPPL 882
>TC232240 weakly similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (38%)
Length = 469
Score = 125 bits (313), Expect = 4e-29
Identities = 68/150 (45%), Positives = 92/150 (61%), Gaps = 6/150 (4%)
Frame = +2
Query: 33 SSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEEL 92
S S+C +PA++N GDSNSDTG + AAF P GI++F + +GR SDGRLIIDFI E
Sbjct: 38 SESECIFPAIFNLGDSNSDTGGLSAAFGQAPPPNGITYFHSPNGRFSDGRLIIDFIAESS 217
Query: 93 EIPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPFYLGMQVAQFIQLQSHI 146
+ YL AYL+S+ SN+ HGANFA G+++RP G+SP L +Q QF ++
Sbjct: 218 GLAYLRAYLDSVASNFTHGANFATAGSTVRPQNTTISQSGYSPISLDVQFVQFSDFKTRS 397
Query: 147 ENLLNQFSSNRTEPPFKSYLPRPEDFSKAL 176
+ + Q FK LP+ E FS+AL
Sbjct: 398 KLVRQQGG------VFKELLPKEEYFSQAL 469
>CD409722
Length = 605
Score = 124 bits (311), Expect = 7e-29
Identities = 61/148 (41%), Positives = 84/148 (56%), Gaps = 6/148 (4%)
Frame = -2
Query: 233 GCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYT 292
GCLP + + LD GCV HN+ A+ FN QL+ +L+ P + TYVD++T
Sbjct: 604 GCLPQNIAKFGTDSSKLDGLGCVSSHNQAAKTFNLQLRALCTKLQGQYPDSNVTYVDIFT 425
Query: 293 AKYELISNASKQGFVNPLEVCCG-----SYYGYRIDCGKKAVVNG-TVYGNPCKNPSQHI 346
K LI+N S+ GF P+ CCG Y R+ CG+ NG T+ C + S++I
Sbjct: 424 IKSSLIANYSRYGFEQPIMACCGYGGPPLNYDSRVSCGETKTFNGTTITAKACNDSSEYI 245
Query: 347 SWDGVHYTQAANKWVAKHIRDGSLSDPP 374
SWDG+HYT+ AN++VA I G SDPP
Sbjct: 244 SWDGIHYTETANQYVASQILTGKYSDPP 161
>TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (94%)
Length = 1541
Score = 112 bits (281), Expect = 2e-25
Identities = 100/354 (28%), Positives = 155/354 (43%), Gaps = 22/354 (6%)
Frame = +1
Query: 40 PAVYNFGDSNSDTG-------VVYAAFAGLQSPGGISFFGNL-SGRASDGRLIIDFITEE 91
PA++ FGDS D G +V A F P G F + +GR +G+L DFI +
Sbjct: 442 PAIFTFGDSIVDVGNNNHQLTIVKANFP----PYGRDFENHFPTGRFCNGKLATDFIADI 609
Query: 92 LEIP-YLSAYLN--SIGSNYRHGANFAAGGASIRPVYG--FSPFYLGMQVAQFIQLQSHI 146
L Y AYLN + G N +GANFA+ + + +S L Q+ + + Q+ +
Sbjct: 610 LGFTSYQPAYLNLKTKGKNLLNGANFASASSGYFELTSKLYSSIPLSKQLEYYKECQTKL 789
Query: 147 ENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLG--------FGLMHTSEEEV 198
Q S++ S A+Y I G +D ++T+++
Sbjct: 790 VEAAGQSSASSI-------------ISDAIYLISAGTSDFVQNYYINPLLNKLYTTDQ-- 924
Query: 199 LRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPH 258
++R ++ +Q LY +GAR + + PIGCLP + N CV
Sbjct: 925 --FSDTLLRCYSNFIQSLYALGARRIGVTSLPPIGCLPAVITLF-----GAHINECVTSL 1083
Query: 259 NKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGS-Y 317
N A FN +L L+ LP D+Y Y+L + S+ GF + CCG+
Sbjct: 1084NSDAINFNEKLNTTSQNLKNMLPGLNLVVFDIYQPLYDLATKPSENGFFEARKACCGTGL 1263
Query: 318 YGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLS 371
I C KK++ GT C N S+++ WDG H ++AANK +A + +S
Sbjct: 1264IEVSILCNKKSI--GT-----CANASEYVFWDGFHPSEAANKVLADELITSGIS 1404
>TC232972 weakly similar to UP|O82681 (O82681) Lanatoside
15'-O-acetylesterase precursor, partial (45%)
Length = 570
Score = 110 bits (275), Expect = 1e-24
Identities = 54/166 (32%), Positives = 84/166 (50%), Gaps = 6/166 (3%)
Frame = +3
Query: 222 RVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLP 281
R F + N P+GC P + + N+D GC+I +N +N LK+ + Q R +L
Sbjct: 6 RTFMVLNLAPVGCYPAFLVEFPHDSSNIDDFGCLISYNNAVLNYNNMLKETLKQTRESLS 185
Query: 282 KAKFTYVDVYTAKYELISNASKQGFVNPLEVCC---GSYYGY--RIDCGKKAVVNGTVY- 335
A YVD ++ EL + + G + CC G Y + ++ CG +NG++
Sbjct: 186 DASVIYVDTHSVLLELFQHPTSHGLQYGTKACCGYGGGDYNFDPKVSCGNTKEINGSIMP 365
Query: 336 GNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
C +P ++SWDG+H T+AANK + I +GS SDPP + C
Sbjct: 366 ATTCNDPYNYVSWDGIHSTEAANKLITFAILNGSFSDPPFIFQEHC 503
>TC212934 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetylesterase
precursor, partial (30%)
Length = 442
Score = 71.6 bits (174), Expect(2) = 3e-24
Identities = 33/55 (60%), Positives = 42/55 (76%)
Frame = +2
Query: 33 SSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDF 87
S S+C++ A++NFGDSNSDTG YAAF G P G+++F +GRASDGRLII F
Sbjct: 71 SHSECNFKAIFNFGDSNSDTGGFYAAFPGESGPYGMTYFKKPAGRASDGRLIIGF 235
Score = 58.2 bits (139), Expect(2) = 3e-24
Identities = 28/70 (40%), Positives = 45/70 (64%), Gaps = 6/70 (8%)
Frame = +3
Query: 84 IIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPFYLGMQVA 137
++DF+ + L +P+LS YL SIGS+Y+HGAN+A +++ V G SPF L +Q+
Sbjct: 225 LLDFLAQALGLPFLSPYLQSIGSDYKHGANYATMASTVLMPNTSLFVTGISPFSLAIQLN 404
Query: 138 QFIQLQSHIE 147
Q Q ++ +E
Sbjct: 405 QMKQFKTKVE 434
>TC205733
Length = 1377
Score = 97.8 bits (242), Expect = 7e-21
Identities = 94/358 (26%), Positives = 151/358 (41%), Gaps = 25/358 (6%)
Frame = +2
Query: 33 SSSKCSYPAVYNFGDSNSDTG----VVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFI 88
+S+K S AV FGDS+ D G + A + Q G G +GR +GR+ DFI
Sbjct: 140 TSAKVS--AVIVFGDSSVDAGNNNFIPTIARSNFQPYGRDFEGGKATGRFCNGRIPTDFI 313
Query: 89 TEELEI-PYLSAYLNSIG--SNYRHGANFAAGGA----------SIRPVYGFSPFYLGMQ 135
+E + PY+ AYL+ S++ G FA+ S+ P++ +Y G Q
Sbjct: 314 SESFGLKPYVPAYLDPKYNISDFASGVTFASAATGYDNATSDVLSVIPLWKQLEYYKGYQ 493
Query: 136 VAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSE 195
+NL ++ + + ++AL+ + +G ND +T
Sbjct: 494 -----------KNLSAYLGESKAK----------DTIAEALHLMSLGTNDF-LENYYTMP 607
Query: 196 EEVLRSIPEMMRNFTYDV-----QVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLD 250
+ P+ +NF + + LY +GAR + P+GCLP E
Sbjct: 608 GRASQFTPQQYQNFLAGIAENFIRSLYGLGARKVSLGGLPPMGCLPL-----ERTTSIAG 772
Query: 251 ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPL 310
N CV +N IA EFN +LK+ +L + LP K + + Y +I GF +
Sbjct: 773 GNDCVARYNNIALEFNNRLKNLTIKLNQELPGLKLVFSNPYYIMLSIIKRPQLYGFESTS 952
Query: 311 EVCCGS---YYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHI 365
CC + GY A G ++ C + S+++ WD H T+ N VAK++
Sbjct: 953 VACCATGMFEMGY-------ACSRGQMFS--CTDASKYVFWDSFHPTEMTNSIVAKYV 1099
>TC205002 similar to UP|Q9XIY6 (Q9XIY6) ESTs AU075322(C11109), partial (14%)
Length = 833
Score = 97.1 bits (240), Expect = 1e-20
Identities = 52/155 (33%), Positives = 78/155 (49%), Gaps = 1/155 (0%)
Frame = +1
Query: 228 NTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTY 287
N GP+GC P + + D GC+ +N ++N+ LK + R +L A Y
Sbjct: 4 NLGPVGCYPGYLVELPHATSDYDEFGCMASYNNAVNDYNKLLKYTLSLTRESLVDASLIY 183
Query: 288 VDVYTAKYELISNAS-KQGFVNPLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHI 346
VD +A EL + + G CCG YG + ++ G + + C P ++
Sbjct: 184 VDTNSALLELFHHPTFYAGLKYSTRTCCG--YGGGVYNFNPKILCGHMLASACDEPHSYV 357
Query: 347 SWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
SWDG+H+T+AANK VA I +GSL DPP P+ + C
Sbjct: 358 SWDGIHFTEAANKIVAHAILNGSLFDPPFPLHEHC 462
>TC203926 weakly similar to PIR|T06696|T06696 lipase homolog T29H11.20 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(54%)
Length = 855
Score = 97.1 bits (240), Expect = 1e-20
Identities = 67/210 (31%), Positives = 100/210 (46%), Gaps = 6/210 (2%)
Frame = +2
Query: 171 DFSKALYTI-DIGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNT 229
DF L+ +IG ND + L T +E +R + + + + +Q L + GA+ +
Sbjct: 32 DFDDTLFWFGEIGVNDYAYTLGSTVSDETIRKLA--ISSVSGALQTLLEKGAKYLVVQGL 205
Query: 230 GPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVD 289
GCL S P + D GCV N + N L+D++ + R+ P+A Y D
Sbjct: 206 PLTGCLTLSMYLAPPD--DRDDIGCVKSVNNQSYYHNLVLQDKLQEFRKQYPQAVILYAD 379
Query: 290 VYTAKYELISNASKQGFVNPLEVCCGS---YYGYRI--DCGKKAVVNGTVYGNPCKNPSQ 344
Y A ++ N SK GF VCCGS Y + + CG N TV C +PSQ
Sbjct: 380 YYDAYRTVMKNPSKFGFKETFNVCCGSGEPPYNFTVFATCG---TPNATV----CSSPSQ 538
Query: 345 HISWDGVHYTQAANKWVAKHIRDGSLSDPP 374
+I+WDGVH T+A K ++ G+ + PP
Sbjct: 539 YINWDGVHLTEAMYKVISSMFLQGNFTQPP 628
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,302,682
Number of Sequences: 63676
Number of extensions: 271827
Number of successful extensions: 1635
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 1490
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1503
length of query: 382
length of database: 12,639,632
effective HSP length: 99
effective length of query: 283
effective length of database: 6,335,708
effective search space: 1793005364
effective search space used: 1793005364
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0400a.7


