

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
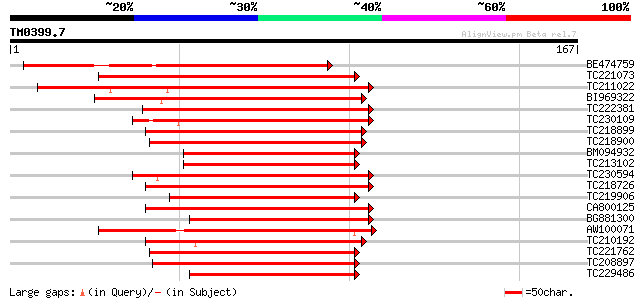
Query= TM0399.7
(167 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE474759 134 3e-32
TC221073 similar to UP|Q93ZL5 (Q93ZL5) AT5g39660/MIJ24_130, part... 130 2e-31
TC211022 similar to UP|O81600 (O81600) H-protein promoter bindin... 129 9e-31
BI969322 104 2e-23
TC222381 similar to UP|Q76KV2 (Q76KV2) DNA binding with one fing... 100 6e-22
TC230109 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP... 99 7e-22
TC218899 similar to UP|Q8L5F0 (Q8L5F0) Dof zinc finger protein, ... 99 1e-21
TC218900 similar to UP|Q9FZA4 (Q9FZA4) F3H9.4 protein, partial (... 99 1e-21
BM094932 98 2e-21
TC213102 homologue to UP|O82155 (O82155) Dof zinc finger protein... 98 2e-21
TC230594 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP... 98 2e-21
TC218726 similar to UP|O82027 (O82027) Dof zinc finger protein (... 98 2e-21
TC219906 homologue to UP|Q9M4G1 (Q9M4G1) Dof zinc finger protein... 98 2e-21
CA800125 similar to PIR|T02370|T023 finger protein BBF1.1 - comm... 98 2e-21
BG881300 similar to GP|21538793|emb dof zinc finger protein {Hor... 97 3e-21
AW100071 similar to GP|9758539|dbj| DNA binding protein-like {Ar... 97 5e-21
TC210192 similar to UP|Q9SSY7 (Q9SSY7) Elicitor-responsive Dof p... 97 5e-21
TC221762 homologue to UP|Q76KV0 (Q76KV0) DNA binding with one fi... 96 6e-21
TC208897 similar to UP|Q9SXG8 (Q9SXG8) Dof zinc finger protein, ... 96 8e-21
TC229486 similar to UP|Q76KV2 (Q76KV2) DNA binding with one fing... 96 8e-21
>BE474759
Length = 301
Score = 134 bits (336), Expect = 3e-32
Identities = 64/91 (70%), Positives = 74/91 (80%)
Frame = +1
Query: 5 QSGQDSEGIKLFGTTITLHGGRETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKF 64
+ ++S+GIKLFG ITL+ G E ++ +KGS ER EK+ +KIIPCPRCKSMETKF
Sbjct: 40 EMAEESQGIKLFGAMITLNRG----EVNKGEKGSEYER-VEKRAEKIIPCPRCKSMETKF 204
Query: 65 CYFNNYNVNQPRHFCKGCQRYWTAGGALRNV 95
CYFNNYNVNQPRHFCK CQRYWTAGGALRNV
Sbjct: 205 CYFNNYNVNQPRHFCKSCQRYWTAGGALRNV 297
>TC221073 similar to UP|Q93ZL5 (Q93ZL5) AT5g39660/MIJ24_130, partial (21%)
Length = 692
Score = 130 bits (328), Expect = 2e-31
Identities = 53/77 (68%), Positives = 65/77 (83%)
Frame = +1
Query: 27 ETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYW 86
+T +++E ++G + +KPDKI+PCPRC SM+TKFCY+NNYNVNQPRHFCK CQRYW
Sbjct: 286 KTSKKTEQEQGENSQDKTLRKPDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKNCQRYW 465
Query: 87 TAGGALRNVPVGAGRRK 103
TAGG +RNVPVGAGRRK
Sbjct: 466 TAGGVMRNVPVGAGRRK 516
>TC211022 similar to UP|O81600 (O81600) H-protein promoter binding factor-2a,
partial (19%)
Length = 578
Score = 129 bits (323), Expect = 9e-31
Identities = 62/112 (55%), Positives = 76/112 (67%), Gaps = 13/112 (11%)
Frame = +1
Query: 9 DSEGIKLFGTTITLHGGRET---RERSEDKKGSGEERDQE----------KKPDKIIPCP 55
D+E K+ GT+ +T E + KG E++Q KKPDK++PCP
Sbjct: 184 DAEETKISGTSPEAIVNPKTPSIEEETAKSKGGKSEKEQGDAANSQEKTLKKPDKVLPCP 363
Query: 56 RCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKMRPS 107
RCKSM+TKFCY+NNYNVNQPR+FCK CQRYWTAGG +RNVPVGAGRRK + S
Sbjct: 364 RCKSMDTKFCYYNNYNVNQPRYFCKACQRYWTAGGTMRNVPVGAGRRKNKNS 519
>BI969322
Length = 650
Score = 104 bits (259), Expect = 2e-23
Identities = 45/83 (54%), Positives = 57/83 (68%), Gaps = 3/83 (3%)
Frame = +3
Query: 26 RETRERSEDKKGSGEERD---QEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGC 82
R+ E +KG E+R Q+ P + CPRC SM TKFCYFNNY+++QPRHFCK C
Sbjct: 156 RDKDMEQEGEKGREEKRQIQQQQPPPQQHQKCPRCDSMNTKFCYFNNYSLSQPRHFCKAC 335
Query: 83 QRYWTAGGALRNVPVGAGRRKMR 105
+RYWT GG RN+PVG G RK++
Sbjct: 336 KRYWTLGGTFRNIPVGGGSRKVK 404
>TC222381 similar to UP|Q76KV2 (Q76KV2) DNA binding with one finger 2
protein, partial (36%)
Length = 485
Score = 99.8 bits (247), Expect = 6e-22
Identities = 40/68 (58%), Positives = 52/68 (75%)
Frame = +1
Query: 40 EERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGA 99
+ER +P++ + CPRC S TKFCY+NNY+++QPR+FCK C+RYWT GG LRNVPVG
Sbjct: 118 QERKPTPQPEQALKCPRCDSTNTKFCYYNNYSLSQPRYFCKSCRRYWTKGGTLRNVPVGG 297
Query: 100 GRRKMRPS 107
G RK + S
Sbjct: 298 GCRKNKRS 321
>TC230109 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP4-like,
partial (27%)
Length = 696
Score = 99.4 bits (246), Expect = 7e-22
Identities = 44/75 (58%), Positives = 54/75 (71%), Gaps = 4/75 (5%)
Frame = +2
Query: 37 GSGEERDQEKKP----DKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGAL 92
GSG D+ +P + + CPRC S+ TKFCY+NNYN++QPRHFCK C+RYWT GG L
Sbjct: 176 GSGSA-DRRLRPHHQNQQALKCPRCDSLNTKFCYYNNYNLSQPRHFCKNCRRYWTKGGVL 352
Query: 93 RNVPVGAGRRKMRPS 107
RNVPVG G RK + S
Sbjct: 353 RNVPVGGGCRKSKRS 397
>TC218899 similar to UP|Q8L5F0 (Q8L5F0) Dof zinc finger protein, partial
(24%)
Length = 692
Score = 98.6 bits (244), Expect = 1e-21
Identities = 37/65 (56%), Positives = 51/65 (77%)
Frame = +1
Query: 41 ERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAG 100
++ Q+ +P + + CPRC S+ TKFCY+NNYN +QPRH+C+ C+R+WT GG LRNVPVG G
Sbjct: 304 KQQQQTQPSEPLKCPRCDSINTKFCYYNNYNKSQPRHYCRACKRHWTKGGTLRNVPVGGG 483
Query: 101 RRKMR 105
R+ R
Sbjct: 484 RKNKR 498
>TC218900 similar to UP|Q9FZA4 (Q9FZA4) F3H9.4 protein, partial (18%)
Length = 693
Score = 98.6 bits (244), Expect = 1e-21
Identities = 37/64 (57%), Positives = 50/64 (77%)
Frame = +1
Query: 42 RDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGR 101
+ Q+ +P + + CPRC S+ TKFCY+NNYN +QPRH+C+ C+R+WT GG LRNVPVG GR
Sbjct: 22 KQQQTQPSEPLKCPRCDSINTKFCYYNNYNKSQPRHYCRACKRHWTKGGTLRNVPVGGGR 201
Query: 102 RKMR 105
+ R
Sbjct: 202 KNKR 213
>BM094932
Length = 412
Score = 98.2 bits (243), Expect = 2e-21
Identities = 38/52 (73%), Positives = 44/52 (84%)
Frame = +2
Query: 52 IPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
+ CPRC S TKFCY+NNYN++QPRHFCK C+RYWT GGALRN+PVG G RK
Sbjct: 23 LQCPRCNSNNTKFCYYNNYNLSQPRHFCKNCRRYWTKGGALRNIPVGGGSRK 178
>TC213102 homologue to UP|O82155 (O82155) Dof zinc finger protein, partial
(34%)
Length = 451
Score = 98.2 bits (243), Expect = 2e-21
Identities = 38/52 (73%), Positives = 44/52 (84%)
Frame = +1
Query: 52 IPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
+ CPRC S TKFCY+NNYN++QPRHFCK C+RYWT GGALRN+PVG G RK
Sbjct: 55 LKCPRCDSTNTKFCYYNNYNLSQPRHFCKNCRRYWTKGGALRNIPVGGGSRK 210
>TC230594 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP4-like,
partial (24%)
Length = 928
Score = 97.8 bits (242), Expect = 2e-21
Identities = 43/75 (57%), Positives = 50/75 (66%), Gaps = 4/75 (5%)
Frame = +3
Query: 37 GSGEER----DQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGAL 92
G G+ R Q I CPRC S+ TKFCY+NNYN++QPRHFCK C+RYWT GG L
Sbjct: 267 GGGDRRLRPHHQNHHQQPPIKCPRCDSLNTKFCYYNNYNLSQPRHFCKNCRRYWTKGGVL 446
Query: 93 RNVPVGAGRRKMRPS 107
RNVP G G RK + S
Sbjct: 447 RNVPFGGGCRKSKRS 491
>TC218726 similar to UP|O82027 (O82027) Dof zinc finger protein (Fragment),
partial (35%)
Length = 1153
Score = 97.8 bits (242), Expect = 2e-21
Identities = 40/67 (59%), Positives = 51/67 (75%)
Frame = +2
Query: 41 ERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAG 100
ER + D+ + CPRC S TKFCY+NNY+++QPR+FCK C+RYWT GG+LRNVPVG G
Sbjct: 188 ERRARPQKDQALNCPRCNSTNTKFCYYNNYSLSQPRYFCKTCRRYWTEGGSLRNVPVGGG 367
Query: 101 RRKMRPS 107
RK + S
Sbjct: 368 SRKNKRS 388
>TC219906 homologue to UP|Q9M4G1 (Q9M4G1) Dof zinc finger protein, partial
(25%)
Length = 1451
Score = 97.8 bits (242), Expect = 2e-21
Identities = 39/56 (69%), Positives = 46/56 (81%)
Frame = +1
Query: 48 PDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
P+ + CPRC S TKFCYFNNY+++QPRHFCK C+RYWT GGALRNVPVG G R+
Sbjct: 376 PEAALKCPRCDSTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGCRR 543
>CA800125 similar to PIR|T02370|T023 finger protein BBF1.1 - common tobacco
(fragment), partial (30%)
Length = 406
Score = 97.8 bits (242), Expect = 2e-21
Identities = 40/67 (59%), Positives = 51/67 (75%)
Frame = +1
Query: 41 ERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAG 100
ER + D+ + CPRC S TKFCY+NNY+++QPR+FCK C+RYWT GG+LRNVPVG G
Sbjct: 205 ERRARPQKDQALNCPRCNSTNTKFCYYNNYSLSQPRYFCKTCRRYWTEGGSLRNVPVGGG 384
Query: 101 RRKMRPS 107
RK + S
Sbjct: 385 SRKNKRS 405
>BG881300 similar to GP|21538793|emb dof zinc finger protein {Hordeum vulgare
subsp. vulgare}, partial (33%)
Length = 394
Score = 97.4 bits (241), Expect = 3e-21
Identities = 39/54 (72%), Positives = 44/54 (81%)
Frame = +3
Query: 54 CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKMRPS 107
CPRC S TKFCY+NNYN+ QPRHFCK C+RYWT GGALRNVP+G G RK + S
Sbjct: 141 CPRCDSSNTKFCYYNNYNLTQPRHFCKTCRRYWTKGGALRNVPIGGGCRKSKSS 302
>AW100071 similar to GP|9758539|dbj| DNA binding protein-like {Arabidopsis
thaliana}, partial (31%)
Length = 339
Score = 96.7 bits (239), Expect = 5e-21
Identities = 42/83 (50%), Positives = 56/83 (66%), Gaps = 1/83 (1%)
Frame = +1
Query: 27 ETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYW 86
E+R S+ + G ++ + +PCPRC S TKFCY+NNYN +QPRHFCK C+RYW
Sbjct: 31 ESRRASKPQSSGGAAPPPPEQEN--LPCPRCDSTNTKFCYYNNYNYSQPRHFCKSCRRYW 204
Query: 87 TAGGALRNVPVGAG-RRKMRPSC 108
T GG LR++PVG G R+ + SC
Sbjct: 205 THGGTLRDIPVGGGSRQNAKRSC 273
>TC210192 similar to UP|Q9SSY7 (Q9SSY7) Elicitor-responsive Dof protein ERDP,
partial (24%)
Length = 1070
Score = 96.7 bits (239), Expect = 5e-21
Identities = 41/66 (62%), Positives = 52/66 (78%), Gaps = 1/66 (1%)
Frame = +3
Query: 41 ERDQEKKPDKIIP-CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGA 99
+R + K ++ P CPRC S TKFCY+NNY+++QPR+FCKGC+RYWT GG+LRNVPVG
Sbjct: 213 QRSRWKPSVEVAPNCPRCASTNTKFCYYNNYSLSQPRYFCKGCRRYWTKGGSLRNVPVGG 392
Query: 100 GRRKMR 105
G RK R
Sbjct: 393 GCRKNR 410
>TC221762 homologue to UP|Q76KV0 (Q76KV0) DNA binding with one finger 4
protein, partial (19%)
Length = 472
Score = 96.3 bits (238), Expect = 6e-21
Identities = 38/62 (61%), Positives = 49/62 (78%)
Frame = +1
Query: 42 RDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGR 101
+D ++ + + CPRC+S TKFCY+NNY+++QPRHFCK C+RYWT GG LRNVPVG G
Sbjct: 115 QDLLQQQQQALKCPRCESSNTKFCYYNNYSLSQPRHFCKACKRYWTRGGTLRNVPVGGGC 294
Query: 102 RK 103
RK
Sbjct: 295 RK 300
>TC208897 similar to UP|Q9SXG8 (Q9SXG8) Dof zinc finger protein, partial
(25%)
Length = 1067
Score = 95.9 bits (237), Expect = 8e-21
Identities = 37/61 (60%), Positives = 49/61 (79%)
Frame = +1
Query: 43 DQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRR 102
+++ +P + + CPRC S TKFCY+NNY++ QPR+FCK C+RYWT GG+LRNVPVG G R
Sbjct: 94 EKKTRPQEQLNCPRCNSTNTKFCYYNNYSLTQPRYFCKTCRRYWTEGGSLRNVPVGGGSR 273
Query: 103 K 103
K
Sbjct: 274 K 276
>TC229486 similar to UP|Q76KV2 (Q76KV2) DNA binding with one finger 2
protein, partial (21%)
Length = 1297
Score = 95.9 bits (237), Expect = 8e-21
Identities = 38/50 (76%), Positives = 42/50 (84%)
Frame = +1
Query: 54 CPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
CPRC S TKFCY+NNYN+ QPRHFCK C+RYWT GGALRNVP+G G RK
Sbjct: 217 CPRCDSSNTKFCYYNNYNLTQPRHFCKTCRRYWTKGGALRNVPIGGGCRK 366
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,703,766
Number of Sequences: 63676
Number of extensions: 141393
Number of successful extensions: 848
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 841
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 844
length of query: 167
length of database: 12,639,632
effective HSP length: 90
effective length of query: 77
effective length of database: 6,908,792
effective search space: 531976984
effective search space used: 531976984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0399.7


