

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0383.9
(421 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
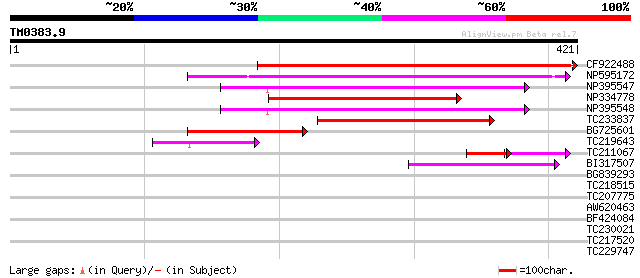
Score E
Sequences producing significant alignments: (bits) Value
CF922488 201 5e-52
NP595172 polyprotein [Glycine max] 168 4e-42
NP395547 reverse transcriptase [Glycine max] 156 1e-38
NP334778 reverse transcriptase [Glycine max] 151 6e-37
NP395548 reverse transcriptase [Glycine max] 143 1e-34
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 134 8e-32
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 98 6e-21
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 61 1e-09
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 35 4e-05
BI317507 42 7e-04
BG839293 38 0.010
TC218515 35 0.063
TC207775 34 0.14
AW620463 29 3.5
BF424084 similar to GP|8778588|gb|A F28C11.4 {Arabidopsis thalia... 28 5.9
TC230021 similar to UP|Q9ARE3 (Q9ARE3) ZF-HD homeobox protein, p... 28 5.9
TC217520 homologue to PIR|T00410|T00410 protein kinase homolog T... 28 7.7
TC229747 28 7.7
>CF922488
Length = 741
Score = 201 bits (511), Expect = 5e-52
Identities = 109/237 (45%), Positives = 147/237 (61%)
Frame = +3
Query: 185 VKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHP 244
V K +GK MCVDY DLN A PK+ +PLP I+ LVD + S +D +SGY+QIK+ P
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 245 SDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVL 304
D +KT F+T +CY+ M FGLKN GATYQR M +F + + +EVY+DDMIVKS
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 305 GSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMK 364
H +L K F RLRK+ +RLNP KC F ++ K L F+ + RGIE++ +K K I EM
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 365 SPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFKSRIK 421
P + K+VQ +GR+ + RF+ P F L KN +W+ +C AF+ RIK
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFE-RIK 710
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 168 bits (426), Expect = 4e-42
Identities = 90/284 (31%), Positives = 154/284 (53%)
Frame = +1
Query: 133 LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKW 192
+ L G PV + ++ I + + ++L I+ P + +++VKK +G W
Sbjct: 1765 IPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSP-FSLPILLVKKKDGSW 1941
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAF 252
R C DY LN K+S+P+P++D+L+D G + S +D SGYHQI + P D +KTAF
Sbjct: 1942 RFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAF 2121
Query: 253 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDL 312
T +Y + MPFGL NA AT+Q LM+++F+ + + + V+ DD+++ S H + L
Sbjct: 2122 RTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHL 2301
Query: 313 TKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEV 372
L++H + KCSFG +LG ++ G+ + K +A+ + +P +VK++
Sbjct: 2302 ESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQL 2481
Query: 373 QHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAF 416
+ +G RF+ + + P L+K+ +F WN+E E AF
Sbjct: 2482 RGFLGLTGYYRRFIKSYANIAGPLTDLLQKD-SFLWNNEAEAAF 2610
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 156 bits (395), Expect = 1e-38
Identities = 87/248 (35%), Positives = 130/248 (52%), Gaps = 18/248 (7%)
Frame = +1
Query: 157 IHQEVNKLLAADFIREIKYPTWMTNVVMVKKANG------------------KWRMCVDY 198
+ +EV KLL A I I +W++ V +V K G +WRMC+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 199 TDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVN 258
LN+A K+ YPLP +D+++ + +D YSGY+QI + P D++KTAF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 259 YCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFAR 318
+ Y+ MPFGL NA T+QR M +F+ V + +EV++DD + +L K R
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 319 LRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGR 378
K N+ LN EKC F +Q G LG I+ RGIE+ +K I ++ P +VK + +G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 379 IAALSRFL 386
+ RF+
Sbjct: 721 VGFYRRFI 744
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 151 bits (381), Expect = 6e-37
Identities = 71/143 (49%), Positives = 96/143 (66%)
Frame = +3
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAF 252
RMCVDY DLN+A PK+++PLP ID L+ + L S +D +SGY+QIKM P D +KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 253 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDL 312
+T +CY+ M FGLKN GATY R M +F+ + + +E YVD+MI KS + H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 313 TKAFARLRKHNMRLNPEKCSFGI 335
F +LRK+ +RLNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 143 bits (361), Expect = 1e-34
Identities = 81/248 (32%), Positives = 132/248 (52%), Gaps = 18/248 (7%)
Frame = +1
Query: 157 IHQEVNKLLAADFIREIKYPTWMTNVVMVKKANG------------------KWRMCVDY 198
+ +EV KLL I I W++ V++V K G W++C+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 199 TDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVN 258
LN+A K+ +PLP +D++++ +G+ +DAY GY+QI + P D++K AF
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 259 YCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFAR 318
+ Y+ +PFGL NA T+Q M +F V +++EV++DD V S + L R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 319 LRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGR 378
+ N+ LN EKC F ++ G LG I+TRGIE++ K I+++ P +VK ++ +G+
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 379 IAALSRFL 386
RF+
Sbjct: 721 ARFYRRFI 744
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 134 bits (337), Expect = 8e-32
Identities = 64/132 (48%), Positives = 91/132 (68%)
Frame = +2
Query: 229 SLIDAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVG 288
S +D +SGY+QI M D +KT F+T + Y+ M FGLKN GATYQR M +F +
Sbjct: 5 SFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMMH 184
Query: 289 RNMEVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTR 348
+ +EVYVDDMI KS + H +L K F RL+K+ ++LNP KC+FG++ GK LGF+++ +
Sbjct: 185 KEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQK 364
Query: 349 GIEINLDKCKAI 360
GIEI+ +K KA+
Sbjct: 365 GIEIDPEKVKAL 400
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 98.2 bits (243), Expect = 6e-21
Identities = 47/89 (52%), Positives = 65/89 (72%)
Frame = -3
Query: 133 LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKW 192
LA+ ++ VTQ +R++ + + + + QEV KL A FIR+I Y T + +VVMVKK NGKW
Sbjct: 271 LAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNGKW 92
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDG 221
R+C DY DLN ACPK++YPLP+ID + DG
Sbjct: 91 RICTDYIDLN*ACPKDAYPLPNIDHMTDG 5
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 60.8 bits (146), Expect = 1e-09
Identities = 31/81 (38%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Frame = +3
Query: 107 MLLGENLDLFAWSHKDMPGIDPNFIC--LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKL 164
+LL + D+FAWS++DMPG+ + + L LNP PV Q R+M I +EV K
Sbjct: 1074 ILLRDYQDIFAWSYQDMPGLSSDIVQHRLPLNPECSPVKQKLRRMKPETSLKIKEEVKK* 1253
Query: 165 LAADFIREIKYPTWMTNVVMV 185
A F+ +YP W+ N+V +
Sbjct: 1254 FDAGFLAVARYPKWVANIVPI 1316
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 35.4 bits (80), Expect(2) = 4e-05
Identities = 16/49 (32%), Positives = 28/49 (56%)
Frame = +1
Query: 368 SVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAF 416
SV +++ G + RF+P+ ++P + +KKN AF W + E+AF
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAF 255
Score = 29.6 bits (65), Expect(2) = 4e-05
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +2
Query: 340 FLGFMITTRGIEINLDKCKAIQEMKSPGSVKEV 372
F GF++ G++++ +K KAIQE P V E+
Sbjct: 23 FSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI 121
>BI317507
Length = 359
Score = 41.6 bits (96), Expect = 7e-04
Identities = 28/112 (25%), Positives = 51/112 (45%)
Frame = -1
Query: 297 DMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDK 356
++++ S SH LT L+K + N +KC F ++LG +I+ + ++ +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 357 CKAIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEW 408
K++ E P +VK V + +F+ G + L KN F+W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKW 18
>BG839293
Length = 781
Score = 37.7 bits (86), Expect = 0.010
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Frame = +1
Query: 107 MLLGENLDLFAWSHKDMPGIDPNFI--CLALNPGIEPVTQ 144
+LL + D+FAWS++DMPG+ + + L LNP PV Q
Sbjct: 610 ILLKDYQDIFAWSYQDMPGLSSDIVQHQLPLNPECSPVKQ 729
>TC218515
Length = 606
Score = 35.0 bits (79), Expect = 0.063
Identities = 13/15 (86%), Positives = 14/15 (92%)
Frame = +3
Query: 258 NYCYQTMPFGLKNAG 272
NYCY+ MPFGLKNAG
Sbjct: 327 NYCYKVMPFGLKNAG 371
>TC207775
Length = 1051
Score = 33.9 bits (76), Expect = 0.14
Identities = 34/108 (31%), Positives = 47/108 (43%), Gaps = 14/108 (12%)
Frame = +1
Query: 156 AIHQEVNKLLAADFIREIKYP---TWMTNVVMV--KKANGKWRMCVDYTD--------LN 202
A + VNK E YP W + + KK G R + +TD +N
Sbjct: 256 AERERVNKAKMESATSESPYPGSGNWEIPIDLFGSKKHAGVSRGVLAFTDESGNIVFRVN 435
Query: 203 KACPK-NSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDK 249
+ P NS PLP KL+ ASGN L S+ ++G + SDE+K
Sbjct: 436 RHPPNPNSSPLPKDKKLLLDASGNTLFSIYRYHNGSWKCYKGNSDENK 579
>AW620463
Length = 398
Score = 29.3 bits (64), Expect = 3.5
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = -1
Query: 49 QRGRPVREKERLGGTLM*RGRCAGREPRHKGRGTS 83
+RGR +R + R GG + RC G R + RG S
Sbjct: 251 ERGRRLRRRRRCGGRGLRGSRCRGAMTRRRVRGRS 147
>BF424084 similar to GP|8778588|gb|A F28C11.4 {Arabidopsis thaliana}, partial
(12%)
Length = 236
Score = 28.5 bits (62), Expect = 5.9
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = -1
Query: 49 QRGRPVREKERLGGTLM*RGRCAGREPRHKGRGTSQQAHADRGDQ 93
+RG E+ L L + R G PRH+GRG +++ GD+
Sbjct: 179 RRGIERVEERNLLALLALQRRLPGPRPRHRGRGEGKESRLADGDR 45
>TC230021 similar to UP|Q9ARE3 (Q9ARE3) ZF-HD homeobox protein, partial (19%)
Length = 986
Score = 28.5 bits (62), Expect = 5.9
Identities = 22/74 (29%), Positives = 35/74 (46%), Gaps = 7/74 (9%)
Frame = -3
Query: 40 PKKGEGVLLQRGRPVREKERLGG-------TLM*RGRCAGREPRHKGRGTSQQAHADRGD 92
P +G+ + RG E+E++G T + G GR GRG A RG+
Sbjct: 243 PSRGD---ITRGERKYEREKMGERGGLGFWTSLKHG--VGRMNCWDGRGKG--ARKKRGE 85
Query: 93 QGAEIWREDTQDRD 106
+G +WR+++ D D
Sbjct: 84 RGGMVWRKESDDDD 43
>TC217520 homologue to PIR|T00410|T00410 protein kinase homolog T13E15.16 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(41%)
Length = 1066
Score = 28.1 bits (61), Expect = 7.7
Identities = 12/38 (31%), Positives = 20/38 (52%)
Frame = -1
Query: 349 GIEINLDKCKAIQEMKSPGSVKEVQHLIGRIAALSRFL 386
G+ L C+ IQE P K VQH++ ++ ++ L
Sbjct: 562 GLYTQLQACEDIQEQAGPPQHKSVQHILKNVSQVAAAL 449
>TC229747
Length = 1327
Score = 28.1 bits (61), Expect = 7.7
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = -1
Query: 298 MIVKSVLGSSHHEDLTKAFARLRKHNMRLNP 328
M + +V + HH++L R+R N RLNP
Sbjct: 181 MQIATVCPTKHHKNLRTRSTRMRNWNRRLNP 89
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,536,885
Number of Sequences: 63676
Number of extensions: 261993
Number of successful extensions: 1059
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1050
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1054
length of query: 421
length of database: 12,639,632
effective HSP length: 100
effective length of query: 321
effective length of database: 6,272,032
effective search space: 2013322272
effective search space used: 2013322272
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0383.9


