

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
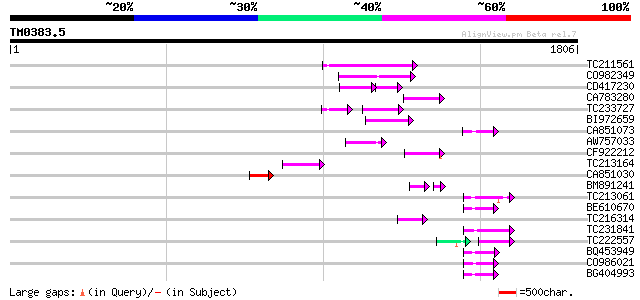
Query= TM0383.5
(1806 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 188 2e-47
CO982349 136 7e-32
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 80 1e-24
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 97 6e-20
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 66 2e-19
BI972659 91 3e-18
CA851073 82 2e-15
AW757033 80 8e-15
CF922212 80 1e-14
TC213164 79 2e-14
CA851030 79 2e-14
BM891241 59 5e-13
TC213061 72 2e-12
BE610670 71 4e-12
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 71 4e-12
TC231841 71 5e-12
TC222557 49 1e-11
BQ453949 69 2e-11
CO986021 68 4e-11
BG404993 67 9e-11
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase (Fragment)
, partial (59%)
Length = 1077
Score = 188 bits (477), Expect = 2e-47
Identities = 103/305 (33%), Positives = 156/305 (50%)
Frame = +1
Query: 995 NAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNLIMN 1054
+A+IA+E +R K + K D KAYD V WNFL + +M F+ +W I
Sbjct: 7 SALIANEVIDEAKRSN---KSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEE 177
Query: 1055 CVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIK 1114
CV + S VL+NG P F P+RGLRQGDPL+P+LF + AE + +++ V +
Sbjct: 178 CVKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYL 357
Query: 1115 ITPRAPMISHLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVP 1174
+ IS L +ADD+I A KE + I++ +E S RINF KS V
Sbjct: 358 VGANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVF-GVT 534
Query: 1175 STRVHELEQLLGVKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGWKEKALSRSG 1234
E L + YLG+P + ++ + + +KL WK + +S G
Sbjct: 535 DQWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGG 714
Query: 1235 REVLIKSIAQAIPSYVMGLFLLPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATPK 1294
R LIKS+ +IP Y F +P ++ +++ ++ +F WGG D +K++WIRW+ L PK
Sbjct: 715 RVTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPK 894
Query: 1295 NMGGL 1299
GG+
Sbjct: 895 ERGGI 909
>CO982349
Length = 795
Score = 136 bits (343), Expect = 7e-32
Identities = 75/248 (30%), Positives = 120/248 (48%)
Frame = +3
Query: 1046 QRWTNLIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKV 1105
Q W I C+ + S +L+N P F P+RGLRQGDPL+P LF + AE + +++ V
Sbjct: 63 QHW---IRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAV 233
Query: 1106 AAGVLHGIKITPRAPMISHLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKS 1165
+ + +S L +ADD+I AT E V+ I++ +E SG +INF +S
Sbjct: 234 DRKRFNSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAES 413
Query: 1166 ELSTSRNVPSTRVHELEQLLGVKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGW 1225
P E + L + YLG+P ++ + + KL W
Sbjct: 414 RFGAIWK-PDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARW 590
Query: 1226 KEKALSRSGREVLIKSIAQAIPSYVMGLFLLPSTLCNQVERMINSFYWGGSADSRKLNWI 1285
K++ +S GR LI +I A+P Y F PS + ++ + F WGG + R++ W+
Sbjct: 591 KQRHISLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRRIAWV 770
Query: 1286 RWDTLATP 1293
+W+T+ P
Sbjct: 771 KWETVCLP 794
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like protein
{Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 80.5 bits (197), Expect(2) = 1e-24
Identities = 40/116 (34%), Positives = 66/116 (56%)
Frame = -3
Query: 1051 LIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVL 1110
L+ C+ST + +L NG+ F P G+RQ DP++PYLF+LC E S +I + +
Sbjct: 680 LVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL* 501
Query: 1111 HGIKITPRAPMISHLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSE 1166
I++ +ISHL F DD IL A ++ V++ ++ + SG ++N DK++
Sbjct: 500 RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTK 333
Score = 52.8 bits (125), Expect(2) = 1e-24
Identities = 27/86 (31%), Positives = 46/86 (53%)
Frame = -2
Query: 1166 ELSTSRNVPSTRVHELEQLLGVKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGW 1225
+ S S+NV ++ LG+++ + GKYLG+P + F F+ +++ K+ W
Sbjct: 333 KFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRFSRW 154
Query: 1226 KEKALSRSGREVLIKSIAQAIPSYVM 1251
K LS GR L K + A+PS++M
Sbjct: 153 KSNLLSMVGRLTLTK*VV*ALPSHIM 76
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase homolog
T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 97.1 bits (240), Expect = 6e-20
Identities = 46/137 (33%), Positives = 73/137 (52%), Gaps = 5/137 (3%)
Frame = +2
Query: 1254 FLLPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATPKNMGGLGFRSFHSFNKALLA 1313
F LP + ++E + F WGG ADSRK+ W+ W T+ PK GGLG + +FN LL
Sbjct: 11 FSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTLLG 190
Query: 1314 KQWWRLINRENSLMVQVIKARYYPKSTPLEANRGTNSSYLWKSVHAAREV-----IQKGS 1368
K W L + +V++++Y E + G+ S WK + +++ +++ +
Sbjct: 191 KWRWDLFYIQQEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPLKRET 370
Query: 1369 IWKVGTGEKIHIWNDQW 1385
IWKVG G++I W D W
Sbjct: 371 IWKVGGGDRIKFWEDLW 421
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 66.2 bits (160), Expect(2) = 2e-19
Identities = 35/99 (35%), Positives = 57/99 (57%)
Frame = -3
Query: 992 ITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNL 1051
I D + E + + +K G G +ALK D+ KA+D ++ +FL S L + G++ + N
Sbjct: 834 IKDCTCVTSEVINMLDKKVFG--GNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNW 661
Query: 1052 IMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLF 1090
I + + + +NG+P F +RG+RQGDPLSP L+
Sbjct: 660 IRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 49.7 bits (117), Expect(2) = 2e-19
Identities = 34/132 (25%), Positives = 59/132 (43%)
Frame = -2
Query: 1123 SHLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELE 1182
SH+ ADD ++ + T L + + Y+ GQ I+ KS++ ++P R+ +
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIG-SIPGQRLRVIS 275
Query: 1183 QLLGVKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGWKEKALSRSGREVLIKSI 1242
LL + Y G+P G+ + V ++I KL WK LS GR L+ S+
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 1243 AQAIPSYVMGLF 1254
+ Y ++
Sbjct: 94 IHGMLLYSFSVY 59
>BI972659
Length = 453
Score = 91.3 bits (225), Expect = 3e-18
Identities = 46/152 (30%), Positives = 82/152 (53%)
Frame = +1
Query: 1134 LARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELEQLLGVKAVEKH 1193
+ A+ + +VL +++G+E SG RIN+ KS+ P+ H+ QLL + ++
Sbjct: 1 VGEASWDNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPNW-AHDAAQLLNCRQLDIP 177
Query: 1194 GKYLGLPTIVGRSKTQVFSFVRERIWKKLKGWKEKALSRSGREVLIKSIAQAIPSYVMGL 1253
YLG+P V S V+ + + KL W +K LS GR LIKS+ A+P Y++
Sbjct: 178 FHYLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSF 357
Query: 1254 FLLPSTLCNQVERMINSFYWGGSADSRKLNWI 1285
F +P + +++ + +F WGG+ +++W+
Sbjct: 358 FKIPQRIVDKLVSLQRTFMWGGNQHHNRISWV 453
>CA851073
Length = 633
Score = 82.4 bits (202), Expect = 2e-15
Identities = 41/116 (35%), Positives = 58/116 (49%), Gaps = 1/116 (0%)
Frame = +1
Query: 1443 THAKDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCN 1502
+H +D ++WK G YS K+ YRI + N + +N++K +WK+K PR
Sbjct: 16 SHLQDTMVWKADPCGVYSTKSAYRILMTCNRH--------VSEANIFKTIWKLKIPPRAA 171
Query: 1503 EIIWRACKGVLPVRMNLKRRGMDL-DTKCPCCGEEDETTTHALLTCSSIRRLWFAS 1557
WR K LP R NL RR + + + +CP CG E E H C R LW+ S
Sbjct: 172 VFSWRLIKDRLPTRHNLLRRNVSIQENECPLCGYEQEEADHLFFNCKMTRGLWWES 339
>AW757033
Length = 441
Score = 80.1 bits (196), Expect = 8e-15
Identities = 46/130 (35%), Positives = 67/130 (51%)
Frame = -2
Query: 1071 RSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFADD 1130
R F P RGLRQGDPL+P LF + AE + +++ VA + +S L +ADD
Sbjct: 437 REFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYADD 258
Query: 1131 SILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELEQLLGVKAV 1190
+I AT E V+ I++G+E SG +INF KS + +VP E + + +
Sbjct: 257 TIFFGEATMENVRVIKTILRGFELASGLKINFAKSRFG-AISVPD*WRKEAAEFMNCSLL 81
Query: 1191 EKHGKYLGLP 1200
YLG+P
Sbjct: 80 SLPFSYLGIP 51
>CF922212
Length = 445
Score = 79.7 bits (195), Expect = 1e-14
Identities = 39/138 (28%), Positives = 66/138 (47%), Gaps = 8/138 (5%)
Frame = -2
Query: 1256 LPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATPKNMGGLGFRSFHSFNKALLAKQ 1315
+P+ + +++ R+ F WGG +K+ W+ WD++ PK GLG R FN ALL K+
Sbjct: 441 VPNKVLDKLVRLQQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKR 262
Query: 1316 WWRLINRENSLMVQVIKARYYPKSTPLEANRGTNSSYLWKSVHAAREVIQKGSIW----- 1370
W L + + L +V+ ++Y E R + S+ W+ + + GS W
Sbjct: 261 RWNLFHHQGELGARVLDSKYKRWRNLDEERRVKSESFWWQEISFITHSTEDGS-WFEKRT 85
Query: 1371 ---KVGTGEKIHIWNDQW 1385
++G K+ W D W
Sbjct: 84 KTGRLGCRAKVRFWEDGW 31
>TC213164
Length = 446
Score = 78.6 bits (192), Expect = 2e-14
Identities = 43/134 (32%), Positives = 70/134 (52%)
Frame = +3
Query: 868 EIMEEPFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGADA 927
+++ F ++EVR + K+ G DG F ++FW I+ D+ + +G
Sbjct: 12 DLLVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFL 191
Query: 928 THINQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFV 987
N +ALIPK+ P +Y+PISL I+K+V K ++ R K L I++E Q+ F+
Sbjct: 192 KRSNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFM 371
Query: 988 PGRLITDNAIIAHE 1001
GR N +IA+E
Sbjct: 372 EGRHTLHNVVIANE 413
>CA851030
Length = 358
Score = 78.6 bits (192), Expect = 2e-14
Identities = 36/75 (48%), Positives = 48/75 (64%)
Frame = +1
Query: 765 KEAEKELENLLKLEEIKWSQRSRATWLAHGDRNTSFFHKKASQRKDRNSIARITDDRGRA 824
K E+ L++LLK EE W QRSRA W D+NT FFH+K QRK NSI +I DD G
Sbjct: 133 KVVEESLDSLLKQEEEYWQQRSRAIWFKERDKNTEFFHRKTLQRKASNSITKIQDDEGNV 312
Query: 825 WTEEEDIEEVLTKYF 839
+ + IE++L K++
Sbjct: 313 CFKPDQIEDILIKFY 357
>BM891241
Length = 407
Score = 58.9 bits (141), Expect(2) = 5e-13
Identities = 23/62 (37%), Positives = 36/62 (57%)
Frame = -2
Query: 1274 GGSADSRKLNWIRWDTLATPKNMGGLGFRSFHSFNKALLAKQWWRLINRENSLMVQVIKA 1333
GG D +K+ W++W+ + PK GGLG + FN ALL K W L + + L ++I +
Sbjct: 406 GGDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
Query: 1334 RY 1335
+Y
Sbjct: 226 KY 221
Score = 35.4 bits (80), Expect(2) = 5e-13
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 5/41 (12%)
Frame = -3
Query: 1351 SYLWKSVH-----AAREVIQKGSIWKVGTGEKIHIWNDQWL 1386
SY W+ + + + + WKVG G+KI+ W D+WL
Sbjct: 174 SYWWRDLRKFYHQSDHSIFHQYMSWKVGCGDKINFWTDKWL 52
>TC213061
Length = 823
Score = 72.4 bits (176), Expect = 2e-12
Identities = 47/174 (27%), Positives = 70/174 (40%), Gaps = 13/174 (7%)
Frame = +2
Query: 1446 KDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNEII 1505
+DQ +W G Y ++ YR+ R G +K++WK+K +
Sbjct: 125 EDQWVWAAESSGSYLARSAYRVIRE--------GIPEEEQDREFKELWKLKVPMKVTMFA 280
Query: 1506 WRACKGVLPVRMNLKRRGMDL-DTKCPCCGEEDETTTHALLTCSSIRRLW---------- 1554
WR K +LP R NL+R+ ++L + CP C DE+ +H CS I +W
Sbjct: 281 WRLLKDILPTRDNLRRKRVELHEYVCPLCRSMDESASHLFFHCSKILPIWWESLSWVKLV 460
Query: 1555 --FASHLNLHLNEELPDNFAGWLEQVLELPNEETMADLFNLIWVIWKRRNAWIF 1606
F H H ++ + + G L L W IWK RN IF
Sbjct: 461 GAFPHHPRHHFHQHSHEVYQG-------LQGNRWKWWWLALTWTIWKHRNDIIF 601
>BE610670
Length = 368
Score = 71.2 bits (173), Expect = 4e-12
Identities = 36/110 (32%), Positives = 55/110 (49%), Gaps = 1/110 (0%)
Frame = -2
Query: 1447 DQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNEIIW 1506
D LIW+ G YS K+ Y + + + S N + K +W +K PR W
Sbjct: 313 DSLIWRADPSGAYSTKSAYNLLKGEG--------SFDNEDSASKIIWNLKIPPRTIAFSW 158
Query: 1507 RACKGVLPVRMNLKRRGMDLDT-KCPCCGEEDETTTHALLTCSSIRRLWF 1555
R K LP + NL+RR ++L + +CP C E+E H + +C+ R LW+
Sbjct: 157 RIFKNRLPTKANLRRRQVELPSYRCPLCDLEEENVGHIMFSCTRTRSLWW 8
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 71.2 bits (173), Expect = 4e-12
Identities = 30/97 (30%), Positives = 51/97 (51%)
Frame = +2
Query: 1234 GREVLIKSIAQAIPSYVMGLFLLPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATP 1293
GR LIKS+ A+P + F +P + +++ + F WGG+ ++ W++W + P
Sbjct: 8 GRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICNP 187
Query: 1294 KNMGGLGFRSFHSFNKALLAKQWWRLINRENSLMVQV 1330
K GGLG + +FN AL + W L + N L ++
Sbjct: 188 KIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>TC231841
Length = 791
Score = 70.9 bits (172), Expect = 5e-12
Identities = 45/166 (27%), Positives = 75/166 (45%), Gaps = 6/166 (3%)
Frame = +3
Query: 1447 DQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNEIIW 1506
DQ +WK GQY+ K+ Y + + G +++++WK+K + W
Sbjct: 63 DQWVWKADPSGQYTAKSAYGVLWGEMFEEQQDG--------VFEELWKLKLPSKITIFAW 218
Query: 1507 RACKGVLPVRMNLKRRGMDL-DTKCPCCGEEDETTTHALLTCSSIRRLWFASHLNLHLNE 1565
R + LP R NL+R+ +++ D +CP C +E+ H CS I +W+ S ++L
Sbjct: 219 RLIRDRLPTRSNLRRKQIEVDDPRCPFCRSAEESAAHLFFHCSRIAPVWWESLSWVNLLG 398
Query: 1566 ELPDNFAGWLEQVLELPNEETMADLFN-----LIWVIWKRRNAWIF 1606
P++ Q + A + L W IWK+RN IF
Sbjct: 399 VFPNHPRQHFLQHIYGVTAGMRASRWKWWWLALTWTIWKQRNNMIF 536
>TC222557
Length = 1002
Score = 48.5 bits (114), Expect(2) = 1e-11
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 6/122 (4%)
Frame = +1
Query: 1492 VWKVKTLPRCNEIIWRACKGVLPVRMNLKRRGMDLDT-KCPCCGEEDETTTHALLTCSSI 1550
+W++K + WR K +P + NL RR + L+ CP C ++E +H C I
Sbjct: 439 LWQLKIPLKATTFAWRLIKERIPTKGNLWRRRVQLNNLMCPFCNRQEEEASHLFFNCPRI 618
Query: 1551 RRLWFAS-HLNLHLNEELPDNFAGWLEQVLELPNEETMAD----LFNLIWVIWKRRNAWI 1605
LW+ S + P +++ L + A L W IW+ RN +
Sbjct: 619 LPLWWESLAWTKTVGVFSPIPRQNYMQHTLISKGKHLQARWKCWWVALTWSIWRHRNRIV 798
Query: 1606 FL 1607
FL
Sbjct: 799 FL 804
Score = 40.8 bits (94), Expect(2) = 1e-11
Identities = 33/122 (27%), Positives = 47/122 (38%), Gaps = 13/122 (10%)
Frame = +3
Query: 1359 AAREVIQKGSIWKVGTGEKIHIWNDQWLPNSSSGRVTTPRNANEGPQRVCELILAG---R 1415
A + Q WKVG G+KI W D WL R PR N Q+ +++ G
Sbjct: 36 AVHGLFQSAIDWKVGCGDKIRFWEDCWLMEQEPLRAKYPRLYNISCQQQKLILVMGFHSA 215
Query: 1416 GMWNESL----------IKATFSGYDASKILQIPLRGTHAKDQLIWKDSGVGQYSVKAGY 1465
+W L ++A S D I D +WK GQ+S ++ Y
Sbjct: 216 NVWEWKLEWRRHLFDNEVQAAASFLDDISWGHI---DRWTLDCWVWKPEPNGQFSTRSAY 386
Query: 1466 RI 1467
R+
Sbjct: 387 RM 392
>BQ453949
Length = 450
Score = 68.9 bits (167), Expect = 2e-11
Identities = 35/116 (30%), Positives = 59/116 (50%), Gaps = 1/116 (0%)
Frame = +3
Query: 1445 AKDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNEI 1504
++D L+W + G YS K+ Y ++++ S T + +K +W +K PR
Sbjct: 24 SRDLLLWGSNSDGSYSTKSAYNFLKNED--------SQTIEDSAFKNIWNLKLPPRACAF 179
Query: 1505 IWRACKGVLPVRMNLKRRGMDL-DTKCPCCGEEDETTTHALLTCSSIRRLWFASHL 1559
R +P ++NL+RR + L + CP C EE+ET H + +C R LW+ + L
Sbjct: 180 S*RIF*NRIPTKVNLRRRHVKLPSSNCPLCDEEEETVGHVMYSCIKTRHLWWETEL 347
>CO986021
Length = 900
Score = 67.8 bits (164), Expect = 4e-11
Identities = 38/113 (33%), Positives = 51/113 (44%), Gaps = 1/113 (0%)
Frame = -1
Query: 1446 KDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNEII 1505
+D ++WK G YS K+ YR+ N HS+ +K +WK+K PR
Sbjct: 786 QDSMLWKADPTGIYSTKSAYRLLLPNNRPG--------QHSSNFKILWKLKIPPRAELFS 631
Query: 1506 WRACKGVLPVRMNLKRRGMDL-DTKCPCCGEEDETTTHALLTCSSIRRLWFAS 1557
WR + L +R NL RR + L D CP CG E H C LW+ S
Sbjct: 630 WRLFRDRLSIRANLLRRHVALQDIMCPLCGNHQEEAGHLFFHCRMTIGLWWES 472
>BG404993
Length = 412
Score = 66.6 bits (161), Expect = 9e-11
Identities = 32/115 (27%), Positives = 50/115 (42%), Gaps = 1/115 (0%)
Frame = -3
Query: 1444 HAKDQLIWKDSGVGQYSVKAGYRIQRSQNNTHPATGPSGTNHSNLWKKVWKVKTLPRCNE 1503
H D WK G YS K Y + + G N ++ ++WK+K P+
Sbjct: 341 HEADSWTWKADPSGNYSTKTAYELLQEAIR--------GDNEDGIFVQLWKLKIPPKAPV 186
Query: 1504 IIWRACKGVLPVRMNLKRRGMDL-DTKCPCCGEEDETTTHALLTCSSIRRLWFAS 1557
WR LP ++NL+RR +++ D+ CP C +E H C+ W+ S
Sbjct: 185 FTWRLINDRLPTKVNLRRRQVEISDSLCPLCNNSEEDAAHLFFNCNKTLPFWWES 21
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.333 0.144 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,692,266
Number of Sequences: 63676
Number of extensions: 1474463
Number of successful extensions: 8944
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 8675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8879
length of query: 1806
length of database: 12,639,632
effective HSP length: 111
effective length of query: 1695
effective length of database: 5,571,596
effective search space: 9443855220
effective search space used: 9443855220
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0383.5


