

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0380.6
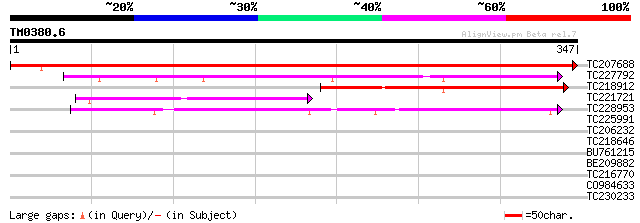
(347 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207688 similar to UP|O65494 (O65494) UDP-galactose transporter... 618 e-177
TC227792 similar to UP|Q9LX35 (Q9LX35) Transporter-like protein,... 143 1e-34
TC218912 similar to GB|AAM60900.1|21536568|AY084311 CMP-sialic a... 121 5e-28
TC221721 similar to GB|AAM60900.1|21536568|AY084311 CMP-sialic a... 114 7e-26
TC228953 similar to UP|Q8VYH1 (Q8VYH1) AT5g65000/MXK3_23, partia... 72 3e-13
TC225991 similar to UP|Q9GSM0 (Q9GSM0) Serine protease K17/F1R2 ... 33 0.25
TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS... 29 2.8
TC218646 weakly similar to UP|Q9FRR1 (Q9FRR1) F22O13.26 (Prenyla... 29 3.6
BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber... 28 4.7
BE209882 weakly similar to SP|P31853|ATPX_ ATP synthase B' chain... 28 6.1
TC216770 similar to UP|Q6K950 (Q6K950) Nucleolar RNA-binding Nop... 28 6.1
CO984633 28 6.1
TC230233 homologue to UP|Q39811 (Q39811) Alpha galactosidase , p... 28 8.0
>TC207688 similar to UP|O65494 (O65494) UDP-galactose transporter-like
protein, partial (98%)
Length = 1401
Score = 618 bits (1594), Expect = e-177
Identities = 317/355 (89%), Positives = 332/355 (93%), Gaps = 8/355 (2%)
Frame = +1
Query: 1 MEYRKIKDEDKVRDSDVE--------SVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFAL 52
MEYRKIKDED+VRD+ VE SV N+ S+GETK+DSHR +V+WKRKS+VT AL
Sbjct: 76 MEYRKIKDEDEVRDAGVEDVGKSFLLSVPDNDLTSEGETKIDSHREKVKWKRKSVVTLAL 255
Query: 53 TILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDNRLTT 112
T+LTSSQ ILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIW K+GV +DNRLTT
Sbjct: 256 TVLTSSQGILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWKKDGVNEDNRLTT 435
Query: 113 TLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEI 172
TLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEI
Sbjct: 436 TLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEI 615
Query: 173 QWAAFILLTAGCTTAQLNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRN 232
Q AAF+LL AGCTTAQLNSNSD VLQTPFQGWVMAI MALLSGFAGVYTEAIIKKRPSRN
Sbjct: 616 QGAAFVLLAAGCTTAQLNSNSDRVLQTPFQGWVMAIVMALLSGFAGVYTEAIIKKRPSRN 795
Query: 233 INVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMK 292
INVQNFWLYVFGM FNAVA+LVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMK
Sbjct: 796 INVQNFWLYVFGMCFNAVAMLVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMK 975
Query: 293 YADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSAGKMQR 347
YADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGT+VVSVAIYLHSAGK+QR
Sbjct: 976 YADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTVVVSVAIYLHSAGKIQR 1140
>TC227792 similar to UP|Q9LX35 (Q9LX35) Transporter-like protein, partial
(84%)
Length = 1781
Score = 143 bits (361), Expect = 1e-34
Identities = 95/322 (29%), Positives = 162/322 (49%), Gaps = 17/322 (5%)
Frame = +1
Query: 34 DSHRTQVQWKRKSMVTFALT---ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKC-- 88
D H++++ K++++ F + +L Q IL+ SK GK+ +S + NF+ E K
Sbjct: 310 DRHKSRISSKQRALNVFLVVGDCVLVGFQPILVYMSKVDGKFNFSPISVNFLTEITKVFF 489
Query: 89 AISLVALGRIWNKEGVTDDNRLTTTLDEV---IVYPIPAALYLVKNLLQYYIFAYVDAPG 145
AI ++ L K G ++T + ++ +PA LY + N L++ + Y +
Sbjct: 490 AIVMLLLQARHQKVGEKPLLSISTFVQAARNNVLLAVPALLYAINNYLKFIMQLYFNPAT 669
Query: 146 YQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNSDHV----LQTPF 201
++L N ++ +L ++I+K+R S IQW A LL G + QL S + L
Sbjct: 670 VKMLSNLKVLVIALLLKVIMKRRFSIIQWEALALLLIGISVNQLRSLPEGTTALGLPVTM 849
Query: 202 QGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVM 261
+ + + A VY E +K + +I +QN +LY +G FN + IL V+
Sbjct: 850 GAYAYTLIFVTVPSLASVYNEYALKSQYDTSIYLQNLFLYGYGAIFNFLGIL----GTVV 1017
Query: 262 NKG-----FFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVF 316
KG G+S T+L+I N+A GI S KYAD I+K YS++VA + T + S
Sbjct: 1018VKGPSSFDILQGHSKATMLLIANNAAQGILSSFFFKYADTILKKYSSTVATIFTGIASAV 1197
Query: 317 LFGFHLSLAFFLGTIVVSVAIY 338
LFG L++ F +G +V ++++
Sbjct: 1198LFGHTLTMNFVIGISIVFISMH 1263
>TC218912 similar to GB|AAM60900.1|21536568|AY084311 CMP-sialic acid
transporter-like protein {Arabidopsis thaliana;} ,
partial (51%)
Length = 958
Score = 121 bits (303), Expect = 5e-28
Identities = 63/157 (40%), Positives = 98/157 (62%), Gaps = 5/157 (3%)
Frame = +2
Query: 191 SNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAV 250
++ D + P QG+++ + A LS AG+YTE ++KK + ++ QN LY FG FN
Sbjct: 47 ASCDSLFSAPIQGYMLGVLSACLSALAGIYTEFLMKKN-NDSLYWQNIQLYTFGTLFNMA 223
Query: 251 AILVQDFDAVMNKG-----FFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSV 305
+L DF G F+GY+ T +++ N +G+ VS +MK+ADNIVKVYSTS+
Sbjct: 224 RLLADDFRGGFENGPWWQRIFNGYTITTWMVVLNLGSTGLLVSWLMKHADNIVKVYSTSM 403
Query: 306 AMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSA 342
AMLLT ++S+FLF F +L FLG I+ ++++++ A
Sbjct: 404 AMLLTMILSLFLFNFKPTLQLFLGIIICMMSLHMYFA 514
>TC221721 similar to GB|AAM60900.1|21536568|AY084311 CMP-sialic acid
transporter-like protein {Arabidopsis thaliana;} ,
partial (38%)
Length = 467
Score = 114 bits (285), Expect = 7e-26
Identities = 60/149 (40%), Positives = 88/149 (58%), Gaps = 4/149 (2%)
Frame = +1
Query: 41 QWKRKSM----VTFALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALG 96
+WK+K M V LT+LTSSQ IL S+ G+Y+Y T F+ E K A+S + L
Sbjct: 28 KWKKKKMQWYFVAALLTVLTSSQGILTTLSQSKGEYKYDYATVPFLAEIFKLAVSSLLLW 207
Query: 97 RIWNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIIS 156
+ K + ++TT V ++PIP+ +YL+ N +Q+ YVD YQIL N I++
Sbjct: 208 KECKKSPLP---KMTTEWKTVSLFPIPSVIYLIHNNVQFATLTYVDTSTYQILGNLKIVT 378
Query: 157 TGVLYRIILKKRLSEIQWAAFILLTAGCT 185
TG+L+R+ L +RLS +QW A +LL G T
Sbjct: 379 TGILFRLFLGRRLSNLQWMAIVLLAVGTT 465
>TC228953 similar to UP|Q8VYH1 (Q8VYH1) AT5g65000/MXK3_23, partial (94%)
Length = 1375
Score = 72.4 bits (176), Expect = 3e-13
Identities = 78/317 (24%), Positives = 141/317 (43%), Gaps = 16/317 (5%)
Frame = +2
Query: 38 TQVQWKRKSMVTFALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLK--CAISLVAL 95
+Q Q + + F +L + SKR + E VT++ E K CA+ +A
Sbjct: 101 SQGQVMNNARIHFFSILLALQYGAQPLISKRFIRREVIVTSSVLTCELAKVICAVFFMA- 277
Query: 96 GRIWNKEG-VTDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNI 154
K+G + + T + + +PAA+Y ++N L + +D+ + +L I
Sbjct: 278 -----KDGSLRKLYKEWTLVGALTASGLPAAIYALQNSLLQISYKNLDSLTFSMLNQTKI 442
Query: 155 ISTGVLYRIILKKRLSEIQWAAFILLTA---------GCTTAQLNSNSDHVLQTPFQGWV 205
T + IL+++ S Q A LL G T N+D +L F G +
Sbjct: 443 FFTALFAYFILRQKQSIEQIGALFLLIVAAVLLSVGEGSTKGSAIGNADQIL---FYGII 613
Query: 206 MAIAMALLSGFAGVYTE--AIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNK 263
+ ++LSG A + + +KK S + ++ V + A + D +A+
Sbjct: 614 PVLVASVLSGLASSLCQWASQVKKHSSYLMTIE--MSIVGSLCLLASTLKSPDGEAMRQH 787
Query: 264 GFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLS 323
GFF+G++ +T++ + +AL GI V +V +A + K + A+L+TA++ G S
Sbjct: 788 GFFYGWTPLTLIPVIFNALGGILVGLVTSHAGGVRKGFVIVSALLITALLQFIFDGKTPS 967
Query: 324 LAFFLG--TIVVSVAIY 338
L L +V S++IY
Sbjct: 968 LYCLLALPLVVTSISIY 1018
>TC225991 similar to UP|Q9GSM0 (Q9GSM0) Serine protease K17/F1R2 (Fragment),
partial (10%)
Length = 901
Score = 32.7 bits (73), Expect = 0.25
Identities = 17/48 (35%), Positives = 25/48 (51%)
Frame = +1
Query: 146 YQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNS 193
Y+I +F + + VLY +I ++ W FI L C+T Q NS S
Sbjct: 745 YKISMSFTLFACSVLYNVIFVCKM*SDGWLKFIGLCNECSTCQSNSLS 888
>TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS-2, partial
(95%)
Length = 960
Score = 29.3 bits (64), Expect = 2.8
Identities = 15/42 (35%), Positives = 22/42 (51%)
Frame = +3
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAIS 91
+ L++L ++ LIV+S R YEYS +E K A S
Sbjct: 171 YELSVLCDAEVALIVFSSRGRLYEYSNNNIRSTIERYKKACS 296
>TC218646 weakly similar to UP|Q9FRR1 (Q9FRR1) F22O13.26 (Prenylated Rab
receptor 4) (At1g08770), partial (62%)
Length = 1127
Score = 28.9 bits (63), Expect = 3.6
Identities = 39/148 (26%), Positives = 70/148 (46%), Gaps = 9/148 (6%)
Frame = +2
Query: 183 GCTTAQLNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNF---- 238
G TT + + V P++ ++ A++ Y +A+I+ R RN++ F
Sbjct: 143 GTTTTTTTTTTSLVTPRPWREFLDLSALSCPYS----YDDAMIRVR--RNLSHFRFNYAA 304
Query: 239 --WLYVF-GMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMKYAD 295
L VF + ++ V+++V F V+ ++ +S L++FN L V V+
Sbjct: 305 ITLLIVFLSLLWHPVSMIV--FLLVLVAWYYLYFSRDVPLVVFNQTLDDRTVLCVLGLL- 475
Query: 296 NIVKVYSTSVAM--LLTAVVSVFLFGFH 321
+V + ST V + LL+ +VSV L G H
Sbjct: 476 TVVSLVSTHVGLNVLLSLIVSVVLVGLH 559
>BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber, partial
(42%)
Length = 450
Score = 28.5 bits (62), Expect = 4.7
Identities = 14/42 (33%), Positives = 23/42 (54%)
Frame = +1
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAIS 91
+ L++L ++ LIV+S R YEY+ + +E K A S
Sbjct: 238 YELSVLCDAEVALIVFSSRGRLYEYANNSVKATIERYKKASS 363
>BE209882 weakly similar to SP|P31853|ATPX_ ATP synthase B' chain
chloroplast precursor (EC 3.6.3.14) (Subunit II).
[Spinach], partial (36%)
Length = 370
Score = 28.1 bits (61), Expect = 6.1
Identities = 17/67 (25%), Positives = 33/67 (48%), Gaps = 8/67 (11%)
Frame = +2
Query: 42 WKRKSMVTFALTILTSSQAILIVWS--------KRAGKYEYSVTTANFMVETLKCAISLV 93
WK K ++ + +VW+ ++AG +++++T MVE C + +V
Sbjct: 53 WKLKLPISKPQMLSLLGGIAPLVWAGPLLTEELEKAGLFDFNLTLPIIMVE---CLLLMV 223
Query: 94 ALGRIWN 100
AL +IW+
Sbjct: 224 ALDKIWS 244
>TC216770 similar to UP|Q6K950 (Q6K950) Nucleolar RNA-binding Nop10p-like
protein, partial (25%)
Length = 647
Score = 28.1 bits (61), Expect = 6.1
Identities = 25/74 (33%), Positives = 38/74 (50%), Gaps = 3/74 (4%)
Frame = -3
Query: 56 TSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKE--GVTDDNRLTTT 113
TSSQ + W+KR K+ S + + TL+ A LV + ++E + +R+ T
Sbjct: 444 TSSQNTVYAWNKRHLKFRASTS-----LNTLEVAAGLVIIQNASSEELSVLNIYHRVKTG 280
Query: 114 LDEVIV-YPIPAAL 126
LDE IV +PA L
Sbjct: 279 LDEQIV*VALPATL 238
>CO984633
Length = 705
Score = 28.1 bits (61), Expect = 6.1
Identities = 21/86 (24%), Positives = 35/86 (40%), Gaps = 8/86 (9%)
Frame = -3
Query: 267 HGYSFITVLMIFNHALSGIAVS--------MVMKYADNIVKVYSTSVAMLLTAVVSVFLF 318
H +S +F A +GI S MV K ++ + M++ ++ +
Sbjct: 574 HAWSLGWDTRLFAPAYAGIVTSGVQYYIQGMVSKIMGPVIVTAFNPLRMIIVTALACIIL 395
Query: 319 GFHLSLAFFLGTIVVSVAIYLHSAGK 344
L L +G IVV + +YL GK
Sbjct: 394 SEQLFLGSIIGAIVVVLGLYLVVWGK 317
>TC230233 homologue to UP|Q39811 (Q39811) Alpha galactosidase , partial (82%)
Length = 1276
Score = 27.7 bits (60), Expect = 8.0
Identities = 14/51 (27%), Positives = 28/51 (54%)
Frame = -1
Query: 84 ETLKCAISLVALGRIWNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQ 134
+ ++CA +AL + ++D + +T LDE++ + I A L L++ Q
Sbjct: 970 DCVECAHKSLALTEVPGFRPMSDQDAVTFALDELLFHKITATLLLLRGPAQ 818
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,599,308
Number of Sequences: 63676
Number of extensions: 167887
Number of successful extensions: 1098
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1090
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1093
length of query: 347
length of database: 12,639,632
effective HSP length: 98
effective length of query: 249
effective length of database: 6,399,384
effective search space: 1593446616
effective search space used: 1593446616
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0380.6


