

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0378b.5
(572 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
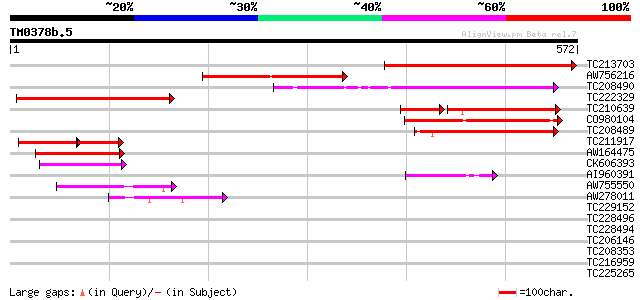
Score E
Sequences producing significant alignments: (bits) Value
TC213703 weakly similar to GB|AAK32900.1|13605829|AF367313 AT5g0... 278 5e-75
AW756216 198 5e-51
TC208490 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T3... 171 6e-43
TC222329 weakly similar to GB|AAK32900.1|13605829|AF367313 AT5g0... 149 3e-36
TC210639 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T3... 114 4e-35
CO980104 143 2e-34
TC208489 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T3... 130 2e-30
TC211917 weakly similar to GB|AAK32900.1|13605829|AF367313 AT5g0... 68 2e-21
AW164475 weakly similar to GP|12003396|gb| Avr9/Cf-9 rapidly eli... 71 1e-12
CK606393 55 6e-08
AI960391 52 7e-07
AW755550 49 6e-06
AW278011 45 1e-04
TC229152 similar to UP|Q6NQ48 (Q6NQ48) At1g34320, partial (32%) 39 0.008
TC228496 33 0.26
TC228494 similar to UP|Q6NQ48 (Q6NQ48) At1g34320, partial (13%) 31 1.3
TC206146 similar to UP|Q8L8M8 (Q8L8M8) Serine-rich protein, part... 30 2.9
TC208353 similar to UP|Q944R5 (Q944R5) At1g14840/F10B6_9, partia... 29 5.0
TC216959 29 6.5
TC225265 homologue to UP|RS14_LUPLU (O22584) 40S ribosomal prote... 28 8.5
>TC213703 weakly similar to GB|AAK32900.1|13605829|AF367313
AT5g04550/T32M21_140 {Arabidopsis thaliana;} , partial
(16%)
Length = 643
Score = 278 bits (711), Expect = 5e-75
Identities = 140/194 (72%), Positives = 158/194 (81%), Gaps = 1/194 (0%)
Frame = +1
Query: 379 NRIRIYSKLSMSNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKML 438
NRIRIYSKLS+ N LKP S TLGDAAL LHYANMIVLIE+M+SSPHL D RDDLY ML
Sbjct: 1 NRIRIYSKLSIKNWLKPVSLTLGDAALALHYANMIVLIERMLSSPHLVDLAARDDLYNML 180
Query: 439 PATIRAVLRHKLKGRKKSKSSW-VYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSF 497
P T+ LR KLK KSKSS ++A AAEWS VLAQIL+WLAPLAH+ + WHS R+F
Sbjct: 181 PTTVTTALRAKLKCHAKSKSSSNAHDANPAAEWSPVLAQILEWLAPLAHNMLSWHSERNF 360
Query: 498 EKEHATLKANILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASARSF 557
EKEH+ AN+LLVQTLYFANQ KTEAAI+DLLVGLNYVCRIDTKVG RD ++C S RSF
Sbjct: 361 EKEHSVFNANVLLVQTLYFANQAKTEAAIIDLLVGLNYVCRIDTKVGTRDTLDCVSTRSF 540
Query: 558 HGVGLRKNGMYSEY 571
+GV LRKNGMY+E+
Sbjct: 541 NGVHLRKNGMYTEF 582
>AW756216
Length = 439
Score = 198 bits (504), Expect = 5e-51
Identities = 106/146 (72%), Positives = 114/146 (77%)
Frame = +1
Query: 195 RQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILERIILVFGNNHLATKQNENESPNTTANN 254
RQEVRNLRDMSPW+RSYDYVVRLL RSLFTILERIILVF N T Q +N+ + ANN
Sbjct: 4 RQEVRNLRDMSPWSRSYDYVVRLLARSLFTILERIILVFAINQPPTVQEQNDYQHMNANN 183
Query: 255 LLRSQSFSVFMHSSVHPSENDLYGFHSGHLGRRLASNSGILVDKKQRKKKQQQALHPPAL 314
LLRS SFSV MHSSVHPSENDLYGF+SG +G R S SG LVDK +RKKKQQQALH PAL
Sbjct: 184 LLRSHSFSV-MHSSVHPSENDLYGFNSGPVGGRPVSKSGFLVDKGRRKKKQQQALHEPAL 360
Query: 315 CRDRLNSESKQFGPIGPFKSCMSVAN 340
R L+SESKQ G I F CMS AN
Sbjct: 361 FRKNLHSESKQLGHIVAFTGCMSAAN 438
>TC208490 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T32M21_140
{Arabidopsis thaliana;} , partial (25%)
Length = 1234
Score = 171 bits (434), Expect = 6e-43
Identities = 111/289 (38%), Positives = 164/289 (56%), Gaps = 2/289 (0%)
Frame = +3
Query: 267 SSVHPSENDLYGFHSGHLGRRLASNSGILVDKKQRKKKQQQALHPPALCRDRLNSESKQ- 325
S + ++N F SG LGR ++ + D K + H + + + ++Q
Sbjct: 18 SGLISAKNRSLKFFSGPLGR---NSKKPVPDNGTNKNSKIWNFHGNSTTTNGKETHTRQS 188
Query: 326 -FGPIGPFKSCMSVANNSPVIDSFVQTNGGSMRLTDSHMENVDKMKRVEKSSLSNRIRIY 384
+ PFK M A++S VIDS N +RL + N K V + +
Sbjct: 189 RLTQVEPFKGFMH-ADSSLVIDSHSSPN--DVRLATQN-PNDPKANLVTPGKEVHHPQ-- 350
Query: 385 SKLSMSNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRA 444
S + RL+P S +LG A+L LHYAN+I++IE++ +SP+L + RDDLY MLP +R+
Sbjct: 351 STFNYLCRLQPPSESLGAASLALHYANVIIMIEKLATSPYLIGVDARDDLYNMLPRRLRS 530
Query: 445 VLRHKLKGRKKSKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATL 504
LR KLK K+ ++ VY+A +A EW+ + IL+WLAPLAH+ +RW S RS+E+
Sbjct: 531 ALRTKLKPYSKAMAAAVYDAGLAEEWTEAMTAILEWLAPLAHNMLRWQSERSYEQHCFVS 710
Query: 505 KANILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECAS 553
+ N+LLVQTLYFA+Q KTEA I +LLVGLNYV R + + ++C S
Sbjct: 711 RTNVLLVQTLYFASQEKTEAIITELLVGLNYVWRYAREFNKKALLDCGS 857
>TC222329 weakly similar to GB|AAK32900.1|13605829|AF367313
AT5g04550/T32M21_140 {Arabidopsis thaliana;} , partial
(23%)
Length = 662
Score = 149 bits (376), Expect = 3e-36
Identities = 75/159 (47%), Positives = 106/159 (66%)
Frame = +3
Query: 8 GSSWLSALWPVSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNS 67
G+SW LW + RK ++++ V+GVL EVA LM K+VNLWQSLSD++V LRE I NS
Sbjct: 186 GASWFRTLWKIRRKDDTNSEKAVIGVLAFEVASLMSKLVNLWQSLSDKQVAKLREEITNS 365
Query: 68 AGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYF 127
G+RKLVSED+++++ L+ E+L+N VA +VAR G++C DP FE+
Sbjct: 366 LGIRKLVSEDENFIVRLISLEMLENMAHVAESVARFGKKCSDPSLKDFENAFDELITFGV 545
Query: 128 QWSGWDYRGKKMERKVKRMEKFVAAMTQLCQELEVLAEV 166
W + KKME+KVKRMEKF++ L QE+E+LA++
Sbjct: 546 DPYRWGFTFKKMEKKVKRMEKFISTNATLYQEMELLADL 662
>TC210639 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T32M21_140
{Arabidopsis thaliana;} , partial (23%)
Length = 701
Score = 114 bits (284), Expect(2) = 4e-35
Identities = 58/118 (49%), Positives = 78/118 (65%), Gaps = 4/118 (3%)
Frame = +3
Query: 442 IRAVLRHKLKGRKK----SKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSF 497
+RA L+ KLK K S SS +Y+ +A EW+ ++ IL+WLAPLAH+ IRW S RS+
Sbjct: 237 VRASLKAKLKPYTKTLASSSSSSIYDPSLAEEWNEAMSSILEWLAPLAHNMIRWQSERSY 416
Query: 498 EKEHATLKANILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASAR 555
E++ + N+LLVQTLYFANQ KTE I +LLVGLNYV + ++ + EC S R
Sbjct: 417 EQQSFISRTNVLLVQTLYFANQEKTEEVITELLVGLNYVWKYGRELNAKALAECGSFR 590
Score = 53.1 bits (126), Expect(2) = 4e-35
Identities = 25/44 (56%), Positives = 32/44 (71%)
Frame = +1
Query: 395 PASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKML 438
P TLG AAL LHYAN+I++IE++ +S HL + RDDLY ML
Sbjct: 97 PPPETLGAAALALHYANVIIVIEKLAASSHLIGLDARDDLYNML 228
>CO980104
Length = 835
Score = 143 bits (361), Expect = 2e-34
Identities = 74/159 (46%), Positives = 106/159 (66%)
Frame = -2
Query: 399 TLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKS 458
TLG AL LHYAN+ ++E+++ PHL E R++LY+MLP ++R L+ KLK K+ +
Sbjct: 699 TLGGCALALHYANVXXVMEKLLRYPHLVGEEARNNLYQMLPTSLRLSLKGKLKTYIKNLA 520
Query: 459 SWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLKANILLVQTLYFAN 518
+Y+A +A +W L IL WLAPLAH+ IRW S R+FE+ + N+LL QTLYFA+
Sbjct: 519 --IYDAPLAHDWKVTLDGILKWLAPLAHNMIRWQSERNFEQHQIVNRTNVLLFQTLYFAD 346
Query: 519 QVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASARSF 557
+ +TE AI LL+GLNY+CR + + V + CAS+ F
Sbjct: 345 KDRTEEAICQLLMGLNYICRYEQQQNV--LLGCASSFDF 235
>TC208489 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T32M21_140
{Arabidopsis thaliana;} , partial (21%)
Length = 841
Score = 130 bits (327), Expect = 2e-30
Identities = 71/148 (47%), Positives = 97/148 (64%), Gaps = 3/148 (2%)
Frame = +2
Query: 409 YANMIVLIEQMVSSPHL---TDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSSWVYNAE 465
YAN V+I S HL + RDDLY MLP +R+ LR KLK K+ ++ VY+A
Sbjct: 2 YAN--VIIHDRRS*QHLHT*LGVDARDDLYNMLPRRLRSALRTKLKPYSKAMAAAVYDAG 175
Query: 466 IAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLKANILLVQTLYFANQVKTEAA 525
+A EW+ + +L+WLAPLAH+ +RW S RS+E+ +AN+LLVQTLYFA+Q KTEA
Sbjct: 176 LADEWTEAMTGMLEWLAPLAHNMLRWQSERSYEQHCFVSRANVLLVQTLYFASQEKTEAI 355
Query: 526 IVDLLVGLNYVCRIDTKVGVRDRVECAS 553
I +LLVGLNYV R ++ + ++C S
Sbjct: 356 ITELLVGLNYVWRYAKELNKKALLDCGS 439
>TC211917 weakly similar to GB|AAK32900.1|13605829|AF367313
AT5g04550/T32M21_140 {Arabidopsis thaliana;} , partial
(11%)
Length = 390
Score = 67.8 bits (164), Expect(2) = 2e-21
Identities = 31/63 (49%), Positives = 45/63 (71%)
Frame = +2
Query: 10 SWLSALWPVSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAG 69
SW +LW RK ++++ VV+ VL E+A LM K+VNLWQSLSD++++ RE I NS+G
Sbjct: 65 SWFRSLWKAPRKHDANSEKVVIEVLAFEIASLMSKLVNLWQSLSDKQIVRFREEITNSSG 244
Query: 70 VRK 72
+K
Sbjct: 245 HKK 253
Score = 53.5 bits (127), Expect(2) = 2e-21
Identities = 22/47 (46%), Positives = 34/47 (71%)
Frame = +3
Query: 69 GVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRF 115
G+RKLVS+DD ++ L+ E+L+N VA +VARL ++C DP++ F
Sbjct: 243 GIRKLVSDDDHFIERLICLEILENMAHVAESVARLAKKCSDPIFKGF 383
>AW164475 weakly similar to GP|12003396|gb| Avr9/Cf-9 rapidly elicited
protein 137 {Nicotiana tabacum}, partial (10%)
Length = 439
Score = 70.9 bits (172), Expect = 1e-12
Identities = 34/90 (37%), Positives = 56/90 (61%)
Frame = +1
Query: 27 KAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVL 86
+A V+G+L + M +++L+ SLSD+E+ LR+ ++NS GV L S+ + +L+ L
Sbjct: 157 RAEVLGILAFDAGKTMCHLISLYHSLSDKEITKLRKEVINSKGVTYLNSQHECFLLNLAA 336
Query: 87 NEVLDNFQSVARTVARLGRRCVDPVYHRFE 116
E L+ + A TV+R GR+C DP RF+
Sbjct: 337 AERLEELDTAADTVSRFGRKCSDPSLSRFD 426
>CK606393
Length = 814
Score = 55.5 bits (132), Expect = 6e-08
Identities = 28/88 (31%), Positives = 50/88 (56%)
Frame = +2
Query: 31 VGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVL 90
+G+L EVA ++K +L +SLS + + L+E ++ V+ LVS+D D L+++V +
Sbjct: 428 IGILAFEVANTIVKGFSLMESLSTKNIKHLKEEVLQLEAVQDLVSKDTDELLKIVGADKR 607
Query: 91 DNFQSVARTVARLGRRCVDPVYHRFEHF 118
D + + V R G R DP +H + +
Sbjct: 608 DELKVFSDEVIRFGNRSKDPQWHNLDRY 691
>AI960391
Length = 417
Score = 52.0 bits (123), Expect = 7e-07
Identities = 32/93 (34%), Positives = 48/93 (51%)
Frame = +1
Query: 400 LGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSS 459
LG+A L+LHYAN+I I + S P + P RD LY LP I++ L ++ K
Sbjct: 52 LGEAGLSLHYANIINQINMIASRPTVLPPNIRDTLYHGLPNNIKSALPSRMXSIDAMKEL 231
Query: 460 WVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWH 492
+ ++ AE + + L WL P A +T + H
Sbjct: 232 SI--TQVKAE----MDKTLQWLNPFATNTTKAH 312
>AW755550
Length = 435
Score = 48.9 bits (115), Expect = 6e-06
Identities = 36/126 (28%), Positives = 57/126 (44%), Gaps = 5/126 (3%)
Frame = +3
Query: 48 LWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRC 107
L+QSL++E + L+ ++ S GV+ LVS D + L+ L + + +R V R G C
Sbjct: 3 LFQSLAEENIQFLKNEVLQSEGVQLLVSNDVEKLITLAEADKREELNVFSREVIRFGNMC 182
Query: 108 VDPVYHRFEHFVRSPAQNYFQWSGWDYRGKK--MERKVKRMEKFVAAM---TQLCQELEV 162
DP +H + YF +D K E K M++F + + +L EL
Sbjct: 183 KDPQWHNLD--------RYFSRLDFDVLDDKRYQEDAEKTMQEFTSLVRNTAELYHELNA 338
Query: 163 LAEVEQ 168
EQ
Sbjct: 339 YERFEQ 356
>AW278011
Length = 391
Score = 44.7 bits (104), Expect = 1e-04
Identities = 34/130 (26%), Positives = 56/130 (42%), Gaps = 10/130 (7%)
Frame = +1
Query: 100 VARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKM-----ERKVKRMEKFVAAMT 154
V R G RC DP +H + YF+ G + +K E ++++ FV
Sbjct: 1 VVRFGNRCKDPQWHNLDR--------YFEKLGSELTPQKQLKEEAEMVMQQLMTFVQYTA 156
Query: 155 QLCQELEVLAEVEQTLRRM-----QANPVLHRGKLPEFQKKVTLQRQEVRNLRDMSPWNR 209
+L EL L +Q RR +N L + ++ Q++ VRNL+ S W++
Sbjct: 157 ELYHELHALDRFDQDYRRKFQEEDNSNATQRGDSLAILRAELKSQKKHVRNLKKKSLWSK 336
Query: 210 SYDYVVRLLV 219
+ V+ LV
Sbjct: 337 ILEEVMEKLV 366
>TC229152 similar to UP|Q6NQ48 (Q6NQ48) At1g34320, partial (32%)
Length = 953
Score = 38.5 bits (88), Expect = 0.008
Identities = 24/94 (25%), Positives = 48/94 (50%), Gaps = 10/94 (10%)
Frame = +1
Query: 453 RKKSKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIR----------WHSVRSFEKEHA 502
R + +S V + A + +IL WL P+A +T + W + S
Sbjct: 16 RSRLQSFQVKEELTVPQIKAEMEKILQWLVPIAANTTKAHHGFGWVGEWANTGSEVNRKP 195
Query: 503 TLKANILLVQTLYFANQVKTEAAIVDLLVGLNYV 536
+ ++L ++TL+ A++ KTEA I++L++ L+++
Sbjct: 196 AGQTDLLRIETLHHADKDKTEAYILELVIWLHHL 297
>TC228496
Length = 762
Score = 33.5 bits (75), Expect = 0.26
Identities = 22/81 (27%), Positives = 38/81 (46%)
Frame = +1
Query: 155 QLCQELEVLAEVEQTLRRMQANPVLHRGKLPEFQKKVTLQRQEVRNLRDMSPWNRSYDYV 214
+L EL+ L ++EQ +R L + ++ +++R+L+ S W RS + V
Sbjct: 34 ELYHELDALDKLEQDFQRKCEEEDQRGDSLALLRAEIKSHMRQIRHLKKKSLWCRSLEEV 213
Query: 215 VRLLVRSLFTILERIILVFGN 235
VR LV + + I GN
Sbjct: 214 VRKLVAIVLFLHLEISNALGN 276
>TC228494 similar to UP|Q6NQ48 (Q6NQ48) At1g34320, partial (13%)
Length = 631
Score = 31.2 bits (69), Expect = 1.3
Identities = 18/66 (27%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Frame = +3
Query: 474 LAQILDWLAPLAHDTIRWHSVRSFEKEHATL-----KANILLVQTLYFANQVKTEAAIVD 528
+ + L WL+P+A +T + H + E A K ++ ++T + A++ K E I++
Sbjct: 195 MEKTLHWLSPMATNTSKAHHGFGWVGEWANTGSEVRKTGVMRIETFHHADKDKVEYYILE 374
Query: 529 LLVGLN 534
LL+ L+
Sbjct: 375 LLLWLH 392
>TC206146 similar to UP|Q8L8M8 (Q8L8M8) Serine-rich protein, partial (42%)
Length = 646
Score = 30.0 bits (66), Expect = 2.9
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = +2
Query: 298 KKQRKKKQQQALHPPALCRDRLNSESKQFGPIGPFKSC 335
KK+RKKK++ L PP+L + + K G K C
Sbjct: 20 KKERKKKKKTLLSPPSLFHRDFSPKRKNGGSFKKIKKC 133
>TC208353 similar to UP|Q944R5 (Q944R5) At1g14840/F10B6_9, partial (25%)
Length = 928
Score = 29.3 bits (64), Expect = 5.0
Identities = 26/101 (25%), Positives = 44/101 (42%), Gaps = 3/101 (2%)
Frame = +2
Query: 56 EVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRC--VDPVYH 113
E ++ E N V+K SE++DY+ ++ + + + V L + C D
Sbjct: 404 ETITTHEESANGTPVKKSKSENEDYVSGML-------YDMLQKEVISLRKACHEKDQTLK 562
Query: 114 RFEHFVRSPAQNYFQWS-GWDYRGKKMERKVKRMEKFVAAM 153
+ + A+ S + +KM R+V MEK VAAM
Sbjct: 563 DKDDAIEMLAKKVDTLSKAMEVEARKMRREVASMEKEVAAM 685
>TC216959
Length = 904
Score = 28.9 bits (63), Expect = 6.5
Identities = 9/22 (40%), Positives = 17/22 (76%)
Frame = -1
Query: 112 YHRFEHFVRSPAQNYFQWSGWD 133
+H FE F+ +P+QN+++W +D
Sbjct: 892 HHYFEAFLCTPSQNHYKWLSFD 827
>TC225265 homologue to UP|RS14_LUPLU (O22584) 40S ribosomal protein S14,
complete
Length = 1024
Score = 28.5 bits (62), Expect = 8.5
Identities = 21/66 (31%), Positives = 31/66 (46%), Gaps = 3/66 (4%)
Frame = +1
Query: 239 ATKQNENESPNTTANNLLRSQSFS---VFMHSSVHPSENDLYGFHSGHLGRRLASNSGIL 295
AT N+ ++P A + LR+ + S + V P +D SG GRRL + GIL
Sbjct: 463 ATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVTPIPSDSTRRKSGRRGRRL*GSRGIL 642
Query: 296 VDKKQR 301
+ R
Sbjct: 643 ISMSNR 660
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,615,112
Number of Sequences: 63676
Number of extensions: 305426
Number of successful extensions: 1501
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1497
length of query: 572
length of database: 12,639,632
effective HSP length: 102
effective length of query: 470
effective length of database: 6,144,680
effective search space: 2887999600
effective search space used: 2887999600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0378b.5


