

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
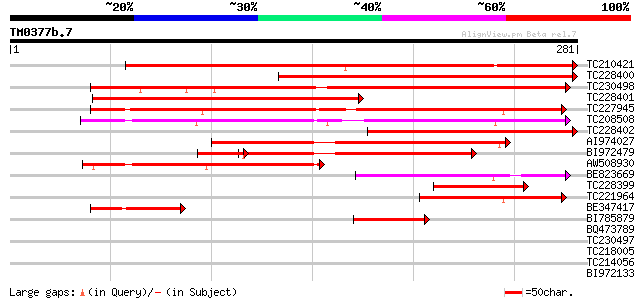
Query= TM0377b.7
(281 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210421 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {A... 351 2e-97
TC228400 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {A... 276 7e-75
TC230498 similar to UP|Q9LPR3 (Q9LPR3) F15H18.4, partial (9%) 239 8e-64
TC228401 weakly similar to GB|AAO24559.1|27808558|BT003127 At5g3... 234 3e-62
TC227945 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {A... 231 3e-61
TC208508 similar to UP|Q9SJI9 (Q9SJI9) Expressed protein, partia... 204 3e-53
TC228402 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {A... 197 6e-51
AI974027 similar to GP|20197959|gb expressed protein {Arabidopsi... 169 1e-42
BI972479 similar to GP|20197959|gb expressed protein {Arabidopsi... 146 6e-39
AW508930 112 2e-25
BE823669 89 2e-18
TC228399 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {A... 89 2e-18
TC221964 weakly similar to GB|AAO24559.1|27808558|BT003127 At5g3... 79 2e-15
BE347417 51 7e-07
BI785879 45 3e-05
BQ473789 similar to GP|26106026|dbj mannose-binding lectin-assoc... 39 0.003
TC230497 33 0.14
TC218005 similar to UP|Q8RVB2 (Q8RVB2) SPY protein, partial (19%) 33 0.14
TC214056 similar to UP|Q9AR81 (Q9AR81) Germin-like protein precu... 30 0.94
BI972133 30 1.2
>TC210421 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Arabidopsis
thaliana;} , partial (47%)
Length = 816
Score = 351 bits (900), Expect = 2e-97
Identities = 170/225 (75%), Positives = 191/225 (84%), Gaps = 1/225 (0%)
Frame = +2
Query: 58 GFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSAQRIPKITYLHIYECEKFSMGIF 117
G PQ+I+ L +VL I+ EDVGLKP+M F S++ +R PKITYLHIYEC++FSMGIF
Sbjct: 8 GLFLSPQNIEMLLSVLGEIKQEDVGLKPEMAFFSSNNPRRTPKITYLHIYECQQFSMGIF 187
Query: 118 CLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKP-TENQALEMRLA 176
CLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPP PTIVKP +E Q +MRLA
Sbjct: 188 CLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPHMPTIVKPSSETQTPDMRLA 367
Query: 177 KVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPF 236
KVKVDADF APC+PSILYP DGGNMH FTAVTACAVLDVLGPPYSD +GRHCTYY N+PF
Sbjct: 368 KVKVDADFNAPCDPSILYPADGGNMHWFTAVTACAVLDVLGPPYSDPDGRHCTYYQNFPF 547
Query: 237 SDISVEGISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGPKIHES 281
S S +G+SIPEEER YEWLQEKE+ E+L+V KMYSGPKI E+
Sbjct: 548 SSYS-DGLSIPEEERTAYEWLQEKEKPENLKVVVKMYSGPKIVEN 679
>TC228400 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Arabidopsis
thaliana;} , partial (41%)
Length = 797
Score = 276 bits (706), Expect = 7e-75
Identities = 126/148 (85%), Positives = 139/148 (93%)
Frame = +3
Query: 134 TVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSIL 193
TVFSKLLFGTMHIKSYDWVVD PPESPT +KP+ENQ EMRLAKVKVDADFTAPCNPSIL
Sbjct: 18 TVFSKLLFGTMHIKSYDWVVDSPPESPTTIKPSENQGPEMRLAKVKVDADFTAPCNPSIL 197
Query: 194 YPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNG 253
YPEDGGN+HCFTAVTACAVLDVLGPPYSD EGRHCTYY+N+PFS+ S +G+SIPEEE+N
Sbjct: 198 YPEDGGNLHCFTAVTACAVLDVLGPPYSDAEGRHCTYYHNFPFSNFSADGLSIPEEEKNA 377
Query: 254 YEWLQEKEQLEDLEVDGKMYSGPKIHES 281
YEWLQE+E+LEDLEV+GKMY+GPKI ES
Sbjct: 378 YEWLQEREELEDLEVNGKMYNGPKIVES 461
>TC230498 similar to UP|Q9LPR3 (Q9LPR3) F15H18.4, partial (9%)
Length = 1137
Score = 239 bits (611), Expect = 8e-64
Identities = 118/250 (47%), Positives = 163/250 (65%), Gaps = 12/250 (4%)
Frame = +2
Query: 41 VQKLFDTCKEVFASGGTGFIPPP-QDIQRLQAVLDAIRPEDVGLKPD-------MPHFRS 92
VQ L+D CK + + G+G PP Q + +L ++LD I+P DVGLK + + F +
Sbjct: 68 VQALYDHCKTILSPSGSGTAPPSSQALMKLSSILDTIQPADVGLKEETADDDRGLGFFGA 247
Query: 93 SSAQRIPK----ITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKS 148
++ R+ + ITY+ I+EC+ F+M IFC P S VIPLH+HPGMTVFSKLL+G++H+K+
Sbjct: 248 NALSRVTRWAQPITYVDIHECDSFTMCIFCFPTSSVIPLHDHPGMTVFSKLLYGSLHVKA 427
Query: 149 YDWVVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVT 208
YDWV E P I++ E ++RLAK+ VD APC+ S+LYP+ GGN+HCFTAVT
Sbjct: 428 YDWV-----EPPCIIESKEPGYAQVRLAKLAVDKVLNAPCDTSVLYPKHGGNLHCFTAVT 592
Query: 209 ACAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWLQEKEQLEDLEV 268
CA+LD+L PPY + EGR CTYY++YP+S S I + E Y WL E E DL +
Sbjct: 593 PCAMLDILTPPYREEEGRRCTYYHDYPYSAFSAANAPIRDGEEEEYAWLTELESPSDLYM 772
Query: 269 DGKMYSGPKI 278
+Y+GP I
Sbjct: 773 RQGVYAGPAI 802
>TC228401 weakly similar to GB|AAO24559.1|27808558|BT003127 At5g39890
{Arabidopsis thaliana;} , partial (41%)
Length = 408
Score = 234 bits (597), Expect = 3e-62
Identities = 106/134 (79%), Positives = 120/134 (89%)
Frame = +1
Query: 42 QKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSAQRIPKI 101
+KL +TCK VFAS GTGF+PP +DI LQ+VLD I+PEDVGL+PDMP+FR+S+ QR+P+I
Sbjct: 7 EKLLETCKVVFASAGTGFVPPHEDIDELQSVLDGIKPEDVGLRPDMPYFRTSATQRVPRI 186
Query: 102 TYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPT 161
TYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSK FGTMHIKSYDWVVD PPESPT
Sbjct: 187 TYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKPTFGTMHIKSYDWVVDSPPESPT 366
Query: 162 IVKPTENQALEMRL 175
+KP+ENQ EMRL
Sbjct: 367 TLKPSENQGPEMRL 408
>TC227945 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Arabidopsis
thaliana;} , partial (47%)
Length = 1772
Score = 231 bits (589), Expect = 3e-61
Identities = 115/238 (48%), Positives = 160/238 (66%), Gaps = 2/238 (0%)
Frame = +3
Query: 41 VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSS-AQRIP 99
+ +LFD+C+E F GT +P PQD++RL +LD ++PEDVGL D+ F+ + +
Sbjct: 486 LHQLFDSCREAFKGPGT--VPSPQDVKRLTHILDNMKPEDVGLSRDLQFFKPGNIVKENQ 659
Query: 100 KITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPES 159
++TY +Y+C+ FS+ IF +P GVIPLHNHP MTVFSKLL G MHIKSYDWV D
Sbjct: 660 RVTYTTVYKCDNFSLCIFFIPEGGVIPLHNHPDMTVFSKLLLGLMHIKSYDWV-DPEASD 836
Query: 160 PTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPP 219
+++P ++RLA +KVD FT+ C+ S+LYP GGN+H FTA+T CAVLDV+GPP
Sbjct: 837 DNMLQPQS----QLRLAMLKVDKVFTSSCDTSVLYPTTGGNIHEFTAITPCAVLDVIGPP 1004
Query: 220 YSDYEGRHCTYYNNYPFSDISVEG-ISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGP 276
YS +GR C+YY ++P++ E I +EE + Y WL+E E E+ E++G Y GP
Sbjct: 1005YSKEDGRDCSYYRDHPYTCFPNERIIGEAKEENDSYTWLEEIEMPENSEMNGVEYLGP 1178
>TC208508 similar to UP|Q9SJI9 (Q9SJI9) Expressed protein, partial (57%)
Length = 1135
Score = 204 bits (520), Expect = 3e-53
Identities = 108/260 (41%), Positives = 156/260 (59%), Gaps = 17/260 (6%)
Frame = +1
Query: 36 KKMPPVQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFR---- 91
+KMP VQKL+DTCK + G + +++++ +LD ++P +VGL+ + R
Sbjct: 256 QKMPIVQKLYDTCKASLSPEGP---ISEEALEKVRTLLDELKPSNVGLEQEAQLVRGWKG 426
Query: 92 -----------SSSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLL 140
+ S Q P I Y+H++EC+KFSMGIFC+ P VIPLHNHPGMTV SKLL
Sbjct: 427 SLNGTNGKKGRNGSYQYPPSIKYIHLHECDKFSMGIFCMSPGSVIPLHNHPGMTVLSKLL 606
Query: 141 FGTMHIKSYDWVVDLP-PESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGG 199
+G++ ++SYDW +DLP P+ P+ +P AK+ D +APCN ++LYP GG
Sbjct: 607 YGSLLVRSYDW-LDLPGPDDPSQARP----------AKLVKDCQMSAPCNTTVLYPSKGG 753
Query: 200 NMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFSDI-SVEGISIPEEERNGYEWLQ 258
N+HCF A+T CA+ DVL PPYS +GRHC+Y+ D+ VE + + + WL+
Sbjct: 754 NIHCFKALTPCALFDVLSPPYSSEDGRHCSYFRKSTRKDLPGVELDQLSGVKPSEITWLE 933
Query: 259 EKEQLEDLEVDGKMYSGPKI 278
E + E+L V +Y GP I
Sbjct: 934 EIQAPENLVVRRGVYKGPTI 993
>TC228402 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Arabidopsis
thaliana;} , partial (31%)
Length = 546
Score = 197 bits (500), Expect = 6e-51
Identities = 87/104 (83%), Positives = 100/104 (95%)
Frame = +1
Query: 178 VKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFS 237
VKVDADFTAPCNPSILYPEDGGN+HCFTAVTACAVLDVLGPPYSD EGRHCTYY+++PFS
Sbjct: 1 VKVDADFTAPCNPSILYPEDGGNLHCFTAVTACAVLDVLGPPYSDAEGRHCTYYHDFPFS 180
Query: 238 DISVEGISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGPKIHES 281
+ SV+G+SIPEEE+N YEWLQE+++LEDLEV+GKMY+GPKI ES
Sbjct: 181 NFSVDGLSIPEEEKNAYEWLQERDELEDLEVNGKMYNGPKIVES 312
>AI974027 similar to GP|20197959|gb expressed protein {Arabidopsis thaliana},
partial (51%)
Length = 431
Score = 169 bits (428), Expect = 1e-42
Identities = 77/152 (50%), Positives = 100/152 (65%), Gaps = 4/152 (2%)
Frame = +3
Query: 101 ITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESP 160
I YLH++EC+ FS+GIFC+PPS +IPLHNHPGMTV SKLL+G+M++KSYDW
Sbjct: 3 IKYLHLHECDSFSIGIFCMPPSSIIPLHNHPGMTVLSKLLYGSMYVKSYDW--------- 155
Query: 161 TIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPY 220
I P N E R AK+ D + TAP ++LYP GGN+HCF A+T CA+ D+L PPY
Sbjct: 156 -IDAPGSNDPSEARPAKLVKDTEMTAPSPTTVLYPTSGGNIHCFRAITPCAIFDILSPPY 332
Query: 221 SDYEGRHCTYYNNYPFSDISV----EGISIPE 248
S GRHCTY+ D+ V G+++ E
Sbjct: 333 SSDHGRHCTYFRRSQRKDLPVNVQLNGVTVSE 428
>BI972479 similar to GP|20197959|gb expressed protein {Arabidopsis thaliana},
partial (51%)
Length = 422
Score = 146 bits (369), Expect(2) = 6e-39
Identities = 65/118 (55%), Positives = 81/118 (68%)
Frame = +2
Query: 114 MGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTENQALEM 173
+GIFC+PPS +IPLHNHPGMTV SKLL+G+M++KSYDW I P N E
Sbjct: 89 IGIFCMPPSSIIPLHNHPGMTVLSKLLYGSMYVKSYDW----------IDAPGSNDPSEA 238
Query: 174 RLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYY 231
R AK+ D + TAP ++LYP GGN+HCF A+T CA+ D+L PPYS GRHCTY+
Sbjct: 239 RPAKLVKDTEMTAPSPTTVLYPTSGGNIHCFRAITPCAIFDILSPPYSSDHGRHCTYF 412
Score = 32.0 bits (71), Expect(2) = 6e-39
Identities = 14/27 (51%), Positives = 19/27 (69%), Gaps = 2/27 (7%)
Frame = +3
Query: 94 SAQRIPKITYLHIYECEKFSM--GIFC 118
S Q +P I YLH++EC+ FS+ IFC
Sbjct: 6 SHQSLPPIKYLHLHECDSFSLICFIFC 86
>AW508930
Length = 444
Score = 112 bits (280), Expect = 2e-25
Identities = 60/130 (46%), Positives = 84/130 (64%), Gaps = 10/130 (7%)
Frame = +1
Query: 37 KMPP-VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSA 95
KMP +Q+L+ CK F+ G + I ++ L+ I+P DVGL+ + R+ S+
Sbjct: 22 KMPYYIQRLYRLCKASFSPNGP---VSEEAIAKVCEKLEKIKPSDVGLEQEAQVVRNWSS 192
Query: 96 Q---------RIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHI 146
Q + P I YLH+YE + FS+GIFC+PPS VIPLHNHPGMTV SKLL+G++++
Sbjct: 193 QMPECNGNHQQPPPIKYLHLYEDDSFSIGIFCMPPSSVIPLHNHPGMTVLSKLLYGSVYV 372
Query: 147 KSYDWVVDLP 156
KSYDW +D P
Sbjct: 373 KSYDW-IDFP 399
>BE823669
Length = 641
Score = 89.0 bits (219), Expect = 2e-18
Identities = 43/111 (38%), Positives = 62/111 (55%), Gaps = 4/111 (3%)
Frame = -3
Query: 172 EMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYY 231
E R AK+ D + TAP ++LYP GGN+H F A+T CA+ DVL PPYS GRHCTY+
Sbjct: 582 EARAAKLVKDTEMTAPTATTVLYPTXGGNIHTFRAITPCAIFDVLSPPYSSEHGRHCTYF 403
Query: 232 NNYPFSD----ISVEGISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGPKI 278
D + + G+++ + WL+E + +D + +Y GP I
Sbjct: 402 RKSQRKDLPGNLQLNGVTVSD-----VTWLEEFQPPDDFVIRRGIYKGPVI 265
>TC228399 similar to GB|AAO24559.1|27808558|BT003127 At5g39890 {Arabidopsis
thaliana;} , partial (17%)
Length = 458
Score = 89.0 bits (219), Expect = 2e-18
Identities = 37/47 (78%), Positives = 44/47 (92%)
Frame = +2
Query: 211 AVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWL 257
AVLDVLGPPYSD EGRHCTYY+++PFS+ SV+G+SIPEEE+N YEWL
Sbjct: 17 AVLDVLGPPYSDAEGRHCTYYHDFPFSNFSVDGLSIPEEEKNAYEWL 157
>TC221964 weakly similar to GB|AAO24559.1|27808558|BT003127 At5g39890
{Arabidopsis thaliana;} , partial (13%)
Length = 824
Score = 79.3 bits (194), Expect = 2e-15
Identities = 37/74 (50%), Positives = 50/74 (67%), Gaps = 1/74 (1%)
Frame = +3
Query: 204 FTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEG-ISIPEEERNGYEWLQEKEQ 262
FTA+T CAVLDV+GPPYS +GR C+YY ++P++ E I +EE + Y WL+E E
Sbjct: 3 FTAITPCAVLDVIGPPYSKEDGRDCSYYRDHPYASFPNERIIGEAKEENDSYAWLEEIEM 182
Query: 263 LEDLEVDGKMYSGP 276
E+ E+DG Y GP
Sbjct: 183 PENSEMDGIEYLGP 224
>BE347417
Length = 480
Score = 50.8 bits (120), Expect = 7e-07
Identities = 22/47 (46%), Positives = 35/47 (73%)
Frame = +1
Query: 41 VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKPDM 87
+Q+LF +C+E F G G +P PQD+ +L +LD+++PEDVGL+ D+
Sbjct: 340 LQELFVSCRETFKGPG-GTVPSPQDVXKLCHILDSMKPEDVGLRSDL 477
>BI785879
Length = 421
Score = 45.4 bits (106), Expect = 3e-05
Identities = 19/38 (50%), Positives = 26/38 (68%)
Frame = +3
Query: 171 LEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVT 208
L R AK+ D +APCN ++L+P GGN+HCF A+T
Sbjct: 306 LAARPAKLMKDCLMSAPCNTTVLHPSKGGNIHCFKALT 419
>BQ473789 similar to GP|26106026|dbj mannose-binding lectin-associated serine
protease {Lethenteron japonicum}, partial (1%)
Length = 421
Score = 38.9 bits (89), Expect = 0.003
Identities = 30/92 (32%), Positives = 42/92 (45%), Gaps = 18/92 (19%)
Frame = +2
Query: 41 VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLKP---------DMPHFR 91
+Q L+D VF+ G +P Q I L+ +LD I DVG+ D
Sbjct: 155 IQVLYDASHAVFSQEG---LPTFQQIHYLKTLLDKIEAIDVGVDESGLCDSPTSDATVDS 325
Query: 92 SSSAQR---------IPKITYLHIYECEKFSM 114
SSS+ +ITY+HI+EC+ FSM
Sbjct: 326 SSSSSNSKGLLCGHGFSEITYIHIHECDYFSM 421
>TC230497
Length = 395
Score = 33.1 bits (74), Expect = 0.14
Identities = 17/44 (38%), Positives = 22/44 (49%)
Frame = +2
Query: 235 PFSDISVEGISIPEEERNGYEWLQEKEQLEDLEVDGKMYSGPKI 278
P+S SV I + E Y WL E E DL + +Y+GP I
Sbjct: 8 PYSAFSVANAPICDGEEEEYAWLTELESPSDLYMRQGVYAGPAI 139
>TC218005 similar to UP|Q8RVB2 (Q8RVB2) SPY protein, partial (19%)
Length = 1395
Score = 33.1 bits (74), Expect = 0.14
Identities = 18/66 (27%), Positives = 34/66 (51%)
Frame = +1
Query: 109 CEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTEN 168
CE MG+ C+ +G + HN G+++ SK+ G + K+ D V L + + + +N
Sbjct: 202 CESLYMGVPCVTMAGSVHAHN-VGVSLLSKVGLGNLIAKNEDEYVKLAVKLASDISALQN 378
Query: 169 QALEMR 174
+ +R
Sbjct: 379 LRMSLR 396
>TC214056 similar to UP|Q9AR81 (Q9AR81) Germin-like protein precursor,
complete
Length = 1020
Score = 30.4 bits (67), Expect = 0.94
Identities = 12/14 (85%), Positives = 12/14 (85%)
Frame = +2
Query: 119 LPPSGVIPLHNHPG 132
L PSGVIPLH HPG
Sbjct: 287 LGPSGVIPLHTHPG 328
>BI972133
Length = 408
Score = 30.0 bits (66), Expect = 1.2
Identities = 13/25 (52%), Positives = 17/25 (68%)
Frame = +3
Query: 1 MGIERTLADRKGRSFCELPRETITS 25
MGI + L++ KGR CEL ET T+
Sbjct: 333 MGIGKNLSETKGRELCELREETNTN 407
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,818,623
Number of Sequences: 63676
Number of extensions: 242191
Number of successful extensions: 1145
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1128
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1129
length of query: 281
length of database: 12,639,632
effective HSP length: 96
effective length of query: 185
effective length of database: 6,526,736
effective search space: 1207446160
effective search space used: 1207446160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0377b.7


