

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
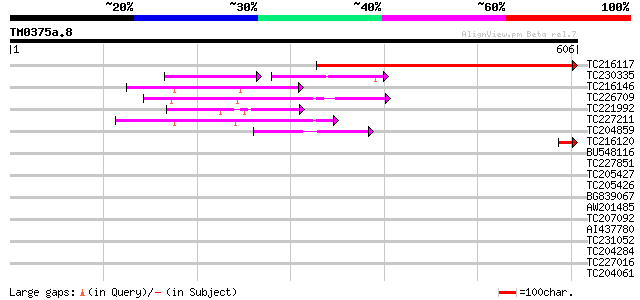
Query= TM0375a.8
(606 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216117 similar to UP|Q8GUG5 (Q8GUG5) Threonine dehydratase/dea... 490 e-139
TC230335 similar to UP|Q8RDT9 (Q8RDT9) Threonine dehydratase , ... 71 2e-25
TC216146 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthas... 66 4e-11
TC226709 similar to UP|CYSK_CITLA (Q43317) Cysteine synthase (B... 58 1e-08
TC221992 homologue to UP|Q81J79 (Q81J79) Cysteine synthase , pa... 57 2e-08
TC227211 UP|Q8W1A0 (Q8W1A0) Cysteine synthase, complete 56 4e-08
TC204859 similar to UP|Q9FS26 (Q9FS26) Plastidic cysteine syntha... 44 2e-04
TC216120 42 6e-04
BU548116 39 0.005
TC227851 homologue to UP|TRP2_ARATH (P25269) Tryptophan synthase... 38 0.011
TC205427 similar to UP|Q94A43 (Q94A43) AT4g36780/C7A10_580, part... 37 0.033
TC205426 similar to UP|Q94A43 (Q94A43) AT4g36780/C7A10_580, part... 37 0.033
BG839067 35 0.096
AW201485 35 0.13
TC207092 similar to UP|Q94ET8 (Q94ET8) LCI5, partial (11%) 34 0.21
AI437780 34 0.21
TC231052 similar to UP|O33678 (O33678) PrsD (ABC transporter ATP... 34 0.21
TC204284 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, ... 33 0.28
TC227016 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.... 33 0.28
TC204061 homologue to UP|Q09083 (Q09083) Hydroxyproline-rich gly... 33 0.28
>TC216117 similar to UP|Q8GUG5 (Q8GUG5) Threonine dehydratase/deaminase
(OMR1), partial (45%)
Length = 1141
Score = 490 bits (1261), Expect = e-139
Identities = 234/278 (84%), Positives = 267/278 (95%)
Frame = +2
Query: 329 VGGFADGVAVKEVGEETFRLCKDLIDGVVLVNRDSICASIKDMFEENRSILEPSGALALA 388
VGGFADGVAVKEVGEETFR+CK+LIDGVVLV+RDSICASIKDMFEE R+ILEP+GALALA
Sbjct: 2 VGGFADGVAVKEVGEETFRICKELIDGVVLVSRDSICASIKDMFEEKRNILEPAGALALA 181
Query: 389 GAEAYCKYYGIKGGNVVAITSGANMNFDKLRVVTELANVGRKQEAVLATVMPEEPGSFKQ 448
GAEAYCK++G++G ++V ITSGANMNFDKLRVVTELANVGRKQEAVLATV+PEEPGSFKQ
Sbjct: 182 GAEAYCKHHGVQGKDIVVITSGANMNFDKLRVVTELANVGRKQEAVLATVLPEEPGSFKQ 361
Query: 449 FCELVGQANITEFTYRFTSSDKAVVLYSVGVHTVSDLSAMKERMKSSHLKTHDLTENDLV 508
FCELVGQ NITEF YR+ S++KAVVLYSVG+HTVS+L AM+ERM+SS LKT++LTE+DLV
Sbjct: 362 FCELVGQMNITEFKYRYNSNEKAVVLYSVGIHTVSELRAMQERMESSQLKTYNLTESDLV 541
Query: 509 KDHLRYMMGGRSDVENEVLCRFIFPERPGALMKFLDSFSPRWNISLFHYRGEGETRANIL 568
KDHLRY+MGGRS+++NEVLCRF FPERPGALMKFLD FSPRWNISLFHYRGEGET AN+L
Sbjct: 542 KDHLRYLMGGRSNIQNEVLCRFTFPERPGALMKFLDPFSPRWNISLFHYRGEGETGANVL 721
Query: 569 VGIQVPRTEMDEFHERANRLGYEYKVVNDDDDFQLLMH 606
VGIQVP++EMDEFH+RAN+LGY+YKVVN+DDDFQLLMH
Sbjct: 722 VGIQVPKSEMDEFHDRANKLGYDYKVVNNDDDFQLLMH 835
>TC230335 similar to UP|Q8RDT9 (Q8RDT9) Threonine dehydratase , partial
(18%)
Length = 908
Score = 71.2 bits (173), Expect(2) = 2e-25
Identities = 45/137 (32%), Positives = 73/137 (52%), Gaps = 12/137 (8%)
Frame = +2
Query: 280 PVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVK 339
P+ GGGLI+GIA K+++P ++I EP A+ A S + + L + ADG+
Sbjct: 347 PISGGGLISGIALAAKSINPAIRIFAAEPKGADDAAQSKAAGRIIRLPETNTIADGLRA- 523
Query: 340 EVGEETFRLCKDLIDGVVLVNRDSICASIKDMFEENRSILEPSGALALAG---------- 389
+G+ T+ + +DL++ ++ V I ++K FE + ++EPSGA+ LA
Sbjct: 524 FLGDFTWPVVQDLVEDIITVEDGEIIKAMKLFFEILKVVVEPSGAIGLAAFLSDTFQKNP 703
Query: 390 AEAYCKYYG--IKGGNV 404
A C + G I GGNV
Sbjct: 704 AWKDCNHIGIVISGGNV 754
Score = 63.5 bits (153), Expect(2) = 2e-25
Identities = 35/104 (33%), Positives = 54/104 (51%)
Frame = +1
Query: 166 LPKELLEKGVICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIG 225
L E KGV+ S+GNHA +ALAAK + + +P P K ++V++ G VV
Sbjct: 7 LNDEDASKGVVTHSSGNHAAALALAAKLRGIPSYIVIPKNAPTCKIENVKRYGGQVVWSE 186
Query: 226 DSYDEAQAYAKKRGIEEGRTFVPAFDHPDVIMGQGTVGMEIMRQ 269
S + A K E G F+ ++ ++ GQGT+ +EI+ Q
Sbjct: 187 ASVQSREEIANKVWQESGAIFIHPYNDGRILSGQGTISLEILEQ 318
>TC216146 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthase, partial
(96%)
Length = 1431
Score = 66.2 bits (160), Expect = 4e-11
Identities = 54/198 (27%), Positives = 91/198 (45%), Gaps = 8/198 (4%)
Frame = +1
Query: 125 SPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLP-KELLEKG---VICSSA 180
+PL K++E G + +K+E +QP S K R AY M+ K L+ G +I ++
Sbjct: 352 TPLVYLNKVTEGCGAYVAVKQEMMQPTASIKDRPAYAMITDAEEKNLITPGKTTLIEPTS 531
Query: 181 GNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGI 240
GN +A A V+ MP T + ++ GA ++L + K +
Sbjct: 532 GNMGISMAFMAAMKGYKMVLTMPSYTSLERRVTMRVFGAELILTDPAKGMGGTVKKAYEL 711
Query: 241 EEGRT---FVPAFDHP-DVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKT 296
E + F +P + + T G EI V + +G GG ++G+ Y+K+
Sbjct: 712 LENTPNAHMLQQFSNPANTQVHFETTGPEIWEDTNGQVDIFVMGIGSGGTVSGVGQYLKS 891
Query: 297 VSPQVKIIGVEPTDANTM 314
+P VKI GVEP+++N +
Sbjct: 892 KNPNVKIYGVEPSESNVL 945
>TC226709 similar to UP|CYSK_CITLA (Q43317) Cysteine synthase
(Beta-pyrazolylalanine synthase) (Beta-PA/CSase)
(L-mimosine synthase) (O-acetylserine sulfhydrylase)
(O-acetylserine (Thiol)-lyase) (CSase) (OAS-TL) ,
complete
Length = 1350
Score = 57.8 bits (138), Expect = 1e-08
Identities = 65/273 (23%), Positives = 115/273 (41%), Gaps = 9/273 (3%)
Frame = +2
Query: 144 KREDLQPVFSFKLRGAYNMMVKLPKELL----EKGVICSSAGNHAQGVALAAKRLNCNAV 199
K E ++P S K R Y+M+V ++ L E +I ++GN G+A A +
Sbjct: 320 KLEMMEPCSSVKDRIGYSMIVDAEEKGLITPGESVLIEPTSGNTGIGLAFMAAAKGYKLI 499
Query: 200 VAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGIEE---GRTFVPAFDHP-DV 255
+ MP + + + GA +VL + A K I + + F++P +
Sbjct: 500 ITMPSSMSLERRTILRAFGAELVLTDPAKGMKGAVQKAEEIRDKTPNSYMLQQFENPANP 679
Query: 256 IMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMA 315
+ T G EI + V A+ +G GG I G Y+K +P +K+ G+EP ++ ++
Sbjct: 680 KVHYETTGPEIWKGSSGKVDALVSGIGTGGTITGAGKYLKEQNPDIKLYGIEPVESPILS 859
Query: 316 LSLHHDQRVILDQVG-GFADGVAVKEVGEETFRLCKDLIDGVVLVNRDSICASIKDMFEE 374
++ +G GF GV L DL+D VV ++ + + K + +
Sbjct: 860 GGKPGPHKI--QGIGAGFIPGV-----------LDVDLLDEVVQISSEEAIETAKLLALK 1000
Query: 375 NRSILEPSGALALAGAEAYCKYYGIKGGNVVAI 407
++ S A A A K G ++A+
Sbjct: 1001EGLLVGISSGAAAAAAIKIAKRPENAGKLIIAV 1099
>TC221992 homologue to UP|Q81J79 (Q81J79) Cysteine synthase , partial (91%)
Length = 893
Score = 57.0 bits (136), Expect = 2e-08
Identities = 47/160 (29%), Positives = 76/160 (47%), Gaps = 12/160 (7%)
Frame = +1
Query: 168 KELLEKG--VICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVL-- 223
K LL++G +I ++GN G+A+ A NA++ MP T + + GA +VL
Sbjct: 223 KGLLKEGDTIIEPTSGNTGIGLAMVAAAKGYNAILVMPETMSIERRNLLRAYGAELVLTP 402
Query: 224 ----IGDSYDEAQAYAKKRGIEEGRTFVPA----FDHPDVIMGQGTVGMEIMRQMVDPVH 275
+G + +A AK+ G F+P +P++ + T G EI+ QM D +
Sbjct: 403 GPEGMGGAIRKATELAKEHGY-----FIPQQFKNQSNPEI--HRLTTGPEIIEQMGDQLD 561
Query: 276 AIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMA 315
A +G GG I G +K +KI VEP D+ ++
Sbjct: 562 AFIAGIGTGGTITGAGEVLKEAYEGIKIYAVEPADSPVLS 681
>TC227211 UP|Q8W1A0 (Q8W1A0) Cysteine synthase, complete
Length = 1256
Score = 56.2 bits (134), Expect = 4e-08
Identities = 59/247 (23%), Positives = 107/247 (42%), Gaps = 9/247 (3%)
Frame = +2
Query: 114 LSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLP-KELLE 172
++ V ++ ++PL KL++ + K E ++P S K R Y+M+ K L+
Sbjct: 110 IAKDVTELIGKTPLVYLNKLADGCVARVAAKLELMEPCSSVKDRIGYSMIADAEEKGLIT 289
Query: 173 KG---VICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYD 229
G +I ++GN G+A A ++ MP + + + GA +VL +
Sbjct: 290 PGKSVLIEPTSGNTGIGLAFMAAARGYKLIITMPASMSLERRIILLAFGAELVLTDPAKG 469
Query: 230 EAQAYAKKRGI---EEGRTFVPAFDHP-DVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGG 285
A K I + F++P + + T G EI + + A +G GG
Sbjct: 470 MKGAVQKAEEILAKTPNAYILQQFENPANPKVHYETTGPEIWKGSDGKIDAFVSGIGTGG 649
Query: 286 LIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVG-GFADGVAVKEVGEE 344
I G Y+K +P +K+IGVEP ++ ++ ++ +G GF GV + +E
Sbjct: 650 TITGAGKYLKEQNPNIKLIGVEPVESPVLSGGKPGPHKI--QGIGAGFIPGVLEVNLLDE 823
Query: 345 TFRLCKD 351
++ D
Sbjct: 824 VIQISSD 844
>TC204859 similar to UP|Q9FS26 (Q9FS26) Plastidic cysteine synthase 1,
partial (63%)
Length = 1092
Score = 43.9 bits (102), Expect = 2e-04
Identities = 31/131 (23%), Positives = 59/131 (44%), Gaps = 3/131 (2%)
Frame = +3
Query: 261 TVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHH 320
T G EI + + +G GG ++G ++K +P++++IGVEP ++N +
Sbjct: 243 TTGPEIWEDTRGKIDILVAGIGTGGTVSGAGQFLKQQNPKIQVIGVEPLESNIL------ 404
Query: 321 DQRVILDQVGGFADGVAVKEVGEETF--RLCKDLIDGVVLVNRDSICASIKDM-FEENRS 377
GG ++ +G L +D++D V+ ++ D + K + +E
Sbjct: 405 --------TGGKPGPHKIQGIGAGFVPRNLDQDVLDEVIAISSDEAVETAKQLALQEGLL 560
Query: 378 ILEPSGALALA 388
+ SGA A A
Sbjct: 561 VGISSGAAAAA 593
>TC216120
Length = 408
Score = 42.4 bits (98), Expect = 6e-04
Identities = 17/20 (85%), Positives = 20/20 (100%)
Frame = +3
Query: 587 RLGYEYKVVNDDDDFQLLMH 606
+LGY+YKVVN+DDDFQLLMH
Sbjct: 3 KLGYDYKVVNNDDDFQLLMH 62
>BU548116
Length = 624
Score = 39.3 bits (90), Expect = 0.005
Identities = 27/111 (24%), Positives = 52/111 (46%), Gaps = 3/111 (2%)
Frame = -2
Query: 281 VGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVKE 340
+G GG G+ ++K +P++++IGVEP ++N + GG ++
Sbjct: 614 IGTGGTXXGVGQFLKQQNPKIQVIGVEPLESNIL--------------TGGKPGPHKIQG 477
Query: 341 VGEETF--RLCKDLIDGVVLVNRDSICASIKDM-FEENRSILEPSGALALA 388
+G L +D++D V+ ++ D + K + +E + SGA A A
Sbjct: 476 IGAGFVPRNLDQDVLDEVIAISSDEAVETAKQLALQEGLLVGISSGAAAAA 324
>TC227851 homologue to UP|TRP2_ARATH (P25269) Tryptophan synthase beta chain
2, chloroplast precursor , partial (83%)
Length = 1557
Score = 38.1 bits (87), Expect = 0.011
Identities = 34/102 (33%), Positives = 48/102 (46%), Gaps = 7/102 (6%)
Frame = +2
Query: 108 TAMANILSAKVYDVAIESPLQLAPKLSERI------GVNLWLKREDLQPVFSFKLRGAYN 161
T +A IL K Y V ESPL A +L+E G +++LKREDL + K+ A
Sbjct: 257 TELAGIL--KDY-VGRESPLYFAERLTEHYKRPNGEGPHIYLKREDLNHTGAHKINNAVA 427
Query: 162 MMVKLPKELLEKGVIC-SSAGNHAQGVALAAKRLNCNAVVAM 202
+ L K L +K +I + AG H A R ++ M
Sbjct: 428 QAL-LAKRLGKKRIIAETGAGQHGVATATVCARFGLECIIYM 550
>TC205427 similar to UP|Q94A43 (Q94A43) AT4g36780/C7A10_580, partial (71%)
Length = 1375
Score = 36.6 bits (83), Expect = 0.033
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 10/70 (14%)
Frame = +1
Query: 14 HPSPPPLLRRHAFHKMPTNVLHPNPRRK----------PFIAATISKPAEITPSPSSSSA 63
+PSP P ++H PT+ P+P R PFI S PA + P S+SA
Sbjct: 514 YPSPVP-----SYHASPTSSSFPSPTRIDGNHPSSFLIPFIRNITSIPANLPPLRISNSA 678
Query: 64 AVGDHLSSPR 73
V LSSPR
Sbjct: 679 PVTPPLSSPR 708
>TC205426 similar to UP|Q94A43 (Q94A43) AT4g36780/C7A10_580, partial (71%)
Length = 1303
Score = 36.6 bits (83), Expect = 0.033
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 10/70 (14%)
Frame = +3
Query: 14 HPSPPPLLRRHAFHKMPTNVLHPNPRRK----------PFIAATISKPAEITPSPSSSSA 63
+PSP P ++H PT+ P+P R PFI S PA + P S+SA
Sbjct: 414 YPSPVP-----SYHASPTSSSFPSPTRIDGNHPSSFLIPFIRNITSIPANLPPLRISNSA 578
Query: 64 AVGDHLSSPR 73
V LSSPR
Sbjct: 579 PVTPPLSSPR 608
>BG839067
Length = 814
Score = 35.0 bits (79), Expect = 0.096
Identities = 32/122 (26%), Positives = 53/122 (43%), Gaps = 10/122 (8%)
Frame = +3
Query: 124 ESPLQLAPKLSERI-------GVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLEKGVI 176
E+PL A +LSE G +++LKREDL S K+ A + + + V
Sbjct: 414 ETPLYHAKRLSEYYKSVNNGKGPDIYLKREDLNHSGSHKMNNALAQAMIAKRMGCKSVVT 593
Query: 177 CSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSV---EKLGATVVLIGDSYDEAQA 233
+ +G H A A +L + V M ++ +V + LGA V + + +A +
Sbjct: 594 ATGSGQHGLATAAACAKLALDCTVFMADKDIHRQYSNVRLIKLLGAQVEAVDGGFTDAAS 773
Query: 234 YA 235
A
Sbjct: 774 EA 779
>AW201485
Length = 411
Score = 34.7 bits (78), Expect = 0.13
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 8/89 (8%)
Frame = +1
Query: 125 SPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLEKGVIC------- 177
+P+ + E + K E +QP FS K R A++M+ K+ +KG+I
Sbjct: 139 TPMVYLNNIVEGCVARIAAKLESMQPCFSIKDRTAFSMI----KDAEDKGLITPGKSVLV 306
Query: 178 -SSAGNHAQGVALAAKRLNCNAVVAMPVT 205
+++GN G+A A +VAMP +
Sbjct: 307 EATSGNTGIGMAFVATLKGYKVIVAMPAS 393
>TC207092 similar to UP|Q94ET8 (Q94ET8) LCI5, partial (11%)
Length = 1015
Score = 33.9 bits (76), Expect = 0.21
Identities = 26/76 (34%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Frame = +1
Query: 7 STTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPF--------IAATI--SKPAEITP 56
S++ P S PP RR A + + P R PF + +TI S P+ +TP
Sbjct: 226 SSSPAPLSSSTPPT-RRSALPESDSTASGSEPTRGPFSTSPSPSPLKSTIETSSPSPMTP 402
Query: 57 SPSSSSAAVGDHLSSP 72
SPS S+ A SSP
Sbjct: 403 SPSPSATAAASSASSP 450
>AI437780
Length = 437
Score = 33.9 bits (76), Expect = 0.21
Identities = 25/74 (33%), Positives = 34/74 (45%)
Frame = +2
Query: 3 ALRFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSS 62
A R +T PSP RR AF + T +L P P + + AA PA+ TPSP ++
Sbjct: 80 AQRSRISTSASTPSPVQF-RRPAFSR--TYILPPTPATRAYAAACWPNPAQPTPSPPPTT 250
Query: 63 AAVGDHLSSPRLRV 76
+ S L V
Sbjct: 251 RSTSGDSSRSELPV 292
>TC231052 similar to UP|O33678 (O33678) PrsD (ABC transporter ATP-binding
protein), partial (4%)
Length = 1171
Score = 33.9 bits (76), Expect = 0.21
Identities = 35/158 (22%), Positives = 70/158 (44%), Gaps = 6/158 (3%)
Frame = +3
Query: 2 AALRFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSS 61
AA+ STT P H PP +A +++ + N + + + ++ P+P+++
Sbjct: 15 AAISSSTTASPPHQDPPAAAPANA---DASHIENTNTAEPVLSKSLVEEVGKVNPAPANA 185
Query: 62 SAAVGDHLSSPRLRVSPDSLQYEPGYL--GAVPDRSRSDGGSDSDAMLTAMANILSAKVY 119
+ ++S + + SPD + + + G P + D L+A+ NIL+ KV
Sbjct: 186 Q----EQIASSKGQESPDQVDPKADEVSTGNTPLDLEAINKMIEDDPLSAIENILTGKV- 350
Query: 120 DVAIESPLQLA----PKLSERIGVNLWLKREDLQPVFS 153
+ ++P + PK+ L + +DL +FS
Sbjct: 351 SICSKTPQSITQSEQPKVQGSPADVLSKELKDLMQIFS 464
>TC204284 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, partial
(60%)
Length = 1202
Score = 33.5 bits (75), Expect = 0.28
Identities = 24/75 (32%), Positives = 33/75 (44%)
Frame = +1
Query: 4 LRFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSA 63
LRF T H + R A + + P P R A IS P TPSP+++SA
Sbjct: 166 LRFMPPTSSHRATRASCSARCASRRPRM*PVKPTPPRSALPAIAISTPP--TPSPAATSA 339
Query: 64 AVGDHLSSPRLRVSP 78
+ SSP ++ P
Sbjct: 340 SPSRRSSSPYIQSRP 384
>TC227016 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.35
{Arabidopsis thaliana;} , partial (25%)
Length = 1286
Score = 33.5 bits (75), Expect = 0.28
Identities = 24/80 (30%), Positives = 36/80 (45%), Gaps = 4/80 (5%)
Frame = +1
Query: 5 RFSTTTHPHHPSPPPLLRRH----AFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSS 60
R P PSPPP L H + +PT PNP P + + + TP S+
Sbjct: 214 RLERAFRPPSPSPPPRLLPHPRDLSLQTLPTP---PNPSSPPSASPSKTSTTSTTPRASA 384
Query: 61 SSAAVGDHLSSPRLRVSPDS 80
SS ++ L++P R + +S
Sbjct: 385 SSPSLAP-LAAPTARSTSNS 441
>TC204061 homologue to UP|Q09083 (Q09083) Hydroxyproline-rich glycoprotein
precursor, partial (28%)
Length = 848
Score = 33.5 bits (75), Expect = 0.28
Identities = 15/33 (45%), Positives = 20/33 (60%)
Frame = +2
Query: 5 RFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPN 37
+F+ T+H HHP+ LL H K PTN HP+
Sbjct: 29 QFTITSHHHHPTSTLLLHHHL--KSPTNTHHPH 121
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,608,999
Number of Sequences: 63676
Number of extensions: 353693
Number of successful extensions: 2954
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 2785
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2926
length of query: 606
length of database: 12,639,632
effective HSP length: 103
effective length of query: 503
effective length of database: 6,081,004
effective search space: 3058745012
effective search space used: 3058745012
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0375a.8


