

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
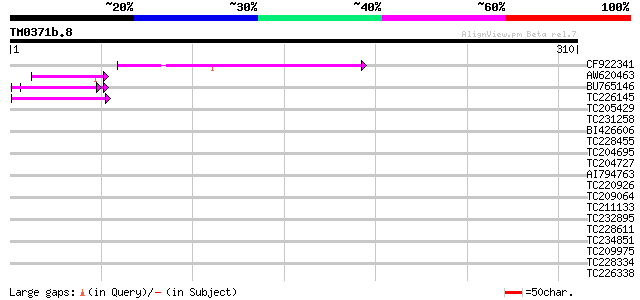
Query= TM0371b.8
(310 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922341 50 2e-06
AW620463 42 4e-04
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 42 5e-04
TC226145 weakly similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine p... 41 8e-04
TC205429 weakly similar to UP|O22600 (O22600) Glycine-rich prote... 40 0.001
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 40 0.001
BI426606 40 0.001
TC228455 similar to UP|Q9M7Y4 (Q9M7Y4) F3E22.8 protein, partial ... 40 0.001
TC204695 similar to UP|Q9LE95 (Q9LE95) Ribosomal protein L1, par... 40 0.002
TC204727 weakly similar to UP|O44358 (O44358) Flagelliform silk ... 40 0.002
AI794763 40 0.002
TC220926 similar to GB|AAF75234.1|8453100|AF233752 short-root pr... 39 0.002
TC209064 similar to UP|P93166 (P93166) SCOF-1, partial (93%) 39 0.002
TC211133 weakly similar to UP|Q9BKV7 (Q9BKV7) Ppg3, partial (4%) 39 0.002
TC232895 weakly similar to PRF|1604369A.0|226743|1604369A sulfat... 39 0.002
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 39 0.002
TC234851 weakly similar to UP|Q9DVW0 (Q9DVW0) PxORF73 peptide, p... 39 0.003
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 39 0.003
TC228334 similar to UP|Q9SW80 (Q9SW80) Bel1-like homeodomain 2 (... 39 0.003
TC226338 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (92%) 39 0.003
>CF922341
Length = 675
Score = 49.7 bits (117), Expect = 2e-06
Identities = 29/140 (20%), Positives = 62/140 (43%), Gaps = 4/140 (2%)
Frame = +1
Query: 60 QWRNLMRSIGNIQQRNEHLQAQLDFYRREQRDDGSRKADSVAEFRPFSED----VESVAI 115
Q+ + + + +N H+ A++ + + ++G R + ++ + + V ++
Sbjct: 232 QYHPQLHLLHSTTSKNPHVMAEMG--KLDHLEEGLRAIEGGEDYAFANLEELFLVPNIIT 405
Query: 116 PDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPR 175
P K L D Y G + PK+HL + KM A + + F + A+ W+T L
Sbjct: 406 PPKFKVLDFDKYKGTTCPKNHLKMYCQKMGAYAKDEELLIHSFQESLTGVAVTWYTNLEP 585
Query: 176 GSISNFRDFSSKFLVQFSAN 195
+ +++D F+ Q+ N
Sbjct: 586 SRVHSWKDLMVAFVRQYQYN 645
>AW620463
Length = 398
Score = 42.0 bits (97), Expect = 4e-04
Identities = 19/44 (43%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Frame = +3
Query: 13 TTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPE--NSPAPEQQP 54
+T P P P PP+PPPPP PS S SP ++P+P P
Sbjct: 207 STTAPPPPPQPPPPLPPPPPPPSSPAS*PSSPSSPSTPSPSSPP 338
Score = 32.0 bits (71), Expect = 0.37
Identities = 14/20 (70%), Positives = 15/20 (75%), Gaps = 2/20 (10%)
Frame = +3
Query: 18 SPSPPP--SPPVPPPPPSPS 35
SPS PP SPP PPP P+PS
Sbjct: 321 SPSSPPMTSPPTPPPGPAPS 380
Score = 27.3 bits (59), Expect = 9.0
Identities = 15/39 (38%), Positives = 16/39 (40%), Gaps = 2/39 (5%)
Frame = +3
Query: 19 PSPPPSPPV--PPPPPSPSQVGSLERSPENSPAPEQQPA 55
P PPPS P P P SPS + P P PA
Sbjct: 258 PPPPPSSPAS*PSSPSSPSTPSPSSPPMTSPPTPPPGPA 374
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 41.6 bits (96), Expect = 5e-04
Identities = 20/49 (40%), Positives = 22/49 (44%)
Frame = -1
Query: 2 PHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 50
P + L P +T P PPP PP PPPPPSP P P P
Sbjct: 248 PPSLXLSPP*KTPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPP 102
Score = 40.8 bits (94), Expect = 8e-04
Identities = 19/48 (39%), Positives = 21/48 (43%)
Frame = -1
Query: 7 LDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
L P + + SP PPP PP PPPPP P P P P P
Sbjct: 407 LPPPPPPSSLPSPPPPPPPPPPPPPPLPPPPPPPPXPPPPPPPPPPPP 264
Score = 40.4 bits (93), Expect = 0.001
Identities = 19/37 (51%), Positives = 19/37 (51%)
Frame = -2
Query: 18 SPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
SP PPP PP PPPPP PS L P P P P
Sbjct: 379 SPPPPPPPPPPPPPPPPS---PLPPPPPPXPPPPPPP 278
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/36 (44%), Positives = 18/36 (49%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PP PP P + L+ P P P P
Sbjct: 285 PPPPPPPPPPPLPPLPXSLSPLKNPPPPPPPPPPPP 178
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/34 (50%), Positives = 18/34 (52%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQ 52
P PPPSPP+PPPPP P P P P Q
Sbjct: 116 PPPPPSPPLPPPPPPPPPPPPPPPPPPPPPPPGQ 15
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/36 (47%), Positives = 18/36 (49%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P SP +P P P
Sbjct: 300 PPPPPPPPPPPPPPPPLPPLPXSLSPLKNPPPPPPP 193
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP PS P P P P
Sbjct: 139 PPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPP 32
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/36 (44%), Positives = 18/36 (49%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP+PPPPP P P P+P P
Sbjct: 181 PPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPP 74
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/45 (37%), Positives = 21/45 (45%)
Frame = -3
Query: 10 PVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P + +++P PPP PP PPPPP P P P P P
Sbjct: 240 PXSLSPLKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPPPP 106
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
PSPPP PP PPPPP P P P P P
Sbjct: 158 PSPPPPPPPPPPPPPPPPPSPPLPPPPPPPPPPPPP 51
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P S P P P P
Sbjct: 151 PPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPP 44
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPPSP P P P P
Sbjct: 363 PPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPP 256
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPPSP P P P P
Sbjct: 133 PPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPP 26
Score = 38.9 bits (89), Expect = 0.003
Identities = 17/37 (45%), Positives = 17/37 (45%)
Frame = -1
Query: 18 SPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
SP PPP PP PPPPP P P P P P
Sbjct: 155 SPPPPPPPPPPPPPPPPPSPPLPPPPPPPPPPPPPPP 45
Score = 38.9 bits (89), Expect = 0.003
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPPSPP PPPPP P P P P P
Sbjct: 170 PPPPPSPPPPPPPPPPPPPPPPPSPPLPPPPPPPPP 63
Score = 38.9 bits (89), Expect = 0.003
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPPSP P P P P
Sbjct: 143 PPPPPPPPPPPPPPSPPLPPPPPPPPPPPPPPPPPP 36
Score = 38.9 bits (89), Expect = 0.003
Identities = 18/46 (39%), Positives = 20/46 (43%)
Frame = -3
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
SP++ P PPP PP PPPPP P P P P P
Sbjct: 228 SPLKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPPPPPLPPP 91
Score = 38.5 bits (88), Expect = 0.004
Identities = 18/35 (51%), Positives = 19/35 (53%), Gaps = 1/35 (2%)
Frame = -1
Query: 21 PPPSPPVPPPPPSPSQVGSLERSPE-NSPAPEQQP 54
PPP PP PPPPP P SL SP +P P P
Sbjct: 296 PPPPPPPPPPPPPPLSPPSLXLSPP*KTPPPPPPP 192
Score = 38.5 bits (88), Expect = 0.004
Identities = 16/36 (44%), Positives = 17/36 (46%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP+PPPPP P P P P P
Sbjct: 353 PPPPPPPPLPPPPPPPPXPPPPPPPPPPPPPPPLSP 246
Score = 38.5 bits (88), Expect = 0.004
Identities = 19/46 (41%), Positives = 20/46 (43%)
Frame = -2
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
SP+ P PPP PP PPPPP PS P P P P
Sbjct: 328 SPLPPPPPPXPPPPPPPP-PPPPPPPSPPPPXXSLPPKKPPPPPPP 194
Score = 38.1 bits (87), Expect = 0.005
Identities = 16/36 (44%), Positives = 17/36 (46%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P P + P P P
Sbjct: 172 PPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPP 65
Score = 38.1 bits (87), Expect = 0.005
Identities = 16/36 (44%), Positives = 16/36 (44%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P P P P P
Sbjct: 148 PPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPP 41
Score = 37.4 bits (85), Expect = 0.009
Identities = 16/37 (43%), Positives = 17/37 (45%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 55
P PPP PP PPPPP P P P P P+
Sbjct: 199 PPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPS 89
Score = 37.4 bits (85), Expect = 0.009
Identities = 16/36 (44%), Positives = 16/36 (44%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P P P P P
Sbjct: 145 PPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPP 38
Score = 37.4 bits (85), Expect = 0.009
Identities = 17/36 (47%), Positives = 18/36 (49%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPP P SL P+ P P P
Sbjct: 292 PPPPPPPPPPPPPSPPPPXXSL--PPKKPPPPPPPP 191
Score = 37.4 bits (85), Expect = 0.009
Identities = 16/36 (44%), Positives = 16/36 (44%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P P P P P
Sbjct: 160 PLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPP 53
Score = 37.0 bits (84), Expect = 0.011
Identities = 15/36 (41%), Positives = 16/36 (43%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPP P P + P P P P
Sbjct: 289 PPPPPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPP 182
Score = 36.6 bits (83), Expect = 0.015
Identities = 13/17 (76%), Positives = 13/17 (76%)
Frame = -2
Query: 18 SPSPPPSPPVPPPPPSP 34
SP PPP PP PPPPP P
Sbjct: 91 SPPPPPPPPPPPPPPPP 41
Score = 35.8 bits (81), Expect = 0.025
Identities = 16/36 (44%), Positives = 16/36 (44%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P P P PP PPPPP P L P P P P
Sbjct: 165 PPPFPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPP 58
Score = 35.4 bits (80), Expect = 0.033
Identities = 16/36 (44%), Positives = 16/36 (44%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP P PPPPP P P SP P P
Sbjct: 175 PPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPP 68
Score = 35.4 bits (80), Expect = 0.033
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSP 44
P PPP PP PPPPP P + SP
Sbjct: 83 PPPPPPPPPPPPPPPPPPPPPGQTSP 6
Score = 35.4 bits (80), Expect = 0.033
Identities = 15/32 (46%), Positives = 15/32 (46%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 50
P PPP PP PPPPP P P P P
Sbjct: 117 PPPPPLPPPPPPPPPPPPPPPPPPPPPPPPPP 22
Score = 35.4 bits (80), Expect = 0.033
Identities = 16/36 (44%), Positives = 16/36 (44%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PP PP PPPPP P SP P P P
Sbjct: 163 PPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPP 56
Score = 34.7 bits (78), Expect = 0.057
Identities = 15/36 (41%), Positives = 16/36 (43%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPP P P P P+P P
Sbjct: 191 PPPPPPPPPPPPSPPPPPPPPPPPPPPPPPSPPLPP 84
Score = 34.7 bits (78), Expect = 0.057
Identities = 15/39 (38%), Positives = 17/39 (43%)
Frame = -1
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
P PPP PP PPP P P P P P P ++
Sbjct: 365 PPPPPPPPPPPPLPPPPPPPPXPPPPPPPPPPPPPPPLS 249
Score = 34.7 bits (78), Expect = 0.057
Identities = 15/36 (41%), Positives = 16/36 (43%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPP P P P P +P
Sbjct: 118 PPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPPGKP 11
Score = 34.3 bits (77), Expect = 0.074
Identities = 15/36 (41%), Positives = 15/36 (41%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPP P P P P P P
Sbjct: 127 PPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPP 20
Score = 33.9 bits (76), Expect = 0.097
Identities = 15/36 (41%), Positives = 15/36 (41%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPP P P P P P P
Sbjct: 193 PPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSP 86
Score = 33.9 bits (76), Expect = 0.097
Identities = 14/32 (43%), Positives = 14/32 (43%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 50
P P P PP PPPPP P P P P
Sbjct: 100 PPPSPPPPPPPPPPPPPPPPPPPPPPPGKPLP 5
Score = 33.9 bits (76), Expect = 0.097
Identities = 14/36 (38%), Positives = 16/36 (43%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP P PPP + + P P P P
Sbjct: 283 PPPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPPPP 176
Score = 33.5 bits (75), Expect = 0.13
Identities = 15/36 (41%), Positives = 15/36 (41%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPP P P P P P
Sbjct: 129 PPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPPPPPP 22
Score = 33.5 bits (75), Expect = 0.13
Identities = 15/36 (41%), Positives = 15/36 (41%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PP PP P P P P P
Sbjct: 135 PPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPPPP 28
Score = 33.5 bits (75), Expect = 0.13
Identities = 15/36 (41%), Positives = 15/36 (41%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPP S P P P P
Sbjct: 280 PPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPPPPP 173
Score = 33.1 bits (74), Expect = 0.16
Identities = 15/36 (41%), Positives = 18/36 (49%)
Frame = -3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP+PP P S S + + P P P P
Sbjct: 276 PPPPPPPPLPPLPXSLSPLKNPPPPPPPPPPPPPPP 169
Score = 32.7 bits (73), Expect = 0.22
Identities = 16/33 (48%), Positives = 16/33 (48%), Gaps = 1/33 (3%)
Frame = -3
Query: 19 PSPPPSPP-VPPPPPSPSQVGSLERSPENSPAP 50
P PPPSPP PPPPP P P P P
Sbjct: 336 PPPPPSPPPPPPPPPPPPPPPPPPPPPPLPPLP 238
Score = 31.6 bits (70), Expect = 0.48
Identities = 17/45 (37%), Positives = 17/45 (37%), Gaps = 8/45 (17%)
Frame = -2
Query: 18 SPSPP--------PSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
SP PP P PP PPPPP P P P P P
Sbjct: 253 SPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPP 119
Score = 31.6 bits (70), Expect = 0.48
Identities = 18/52 (34%), Positives = 18/52 (34%), Gaps = 16/52 (30%)
Frame = -2
Query: 19 PSPPPS----------------PPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPPS PP PPPPP P L P P P P
Sbjct: 268 PPPPPSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPP 113
Score = 30.0 bits (66), Expect = 1.4
Identities = 15/35 (42%), Positives = 16/35 (44%), Gaps = 3/35 (8%)
Frame = -2
Query: 19 PSPPPSP---PVPPPPPSPSQVGSLERSPENSPAP 50
P PPPSP P PP PP P P + P P
Sbjct: 343 PPPPPSPLPPPPPPXPPPPPPPPPPPPPPPSPPPP 239
Score = 27.7 bits (60), Expect = 6.9
Identities = 16/53 (30%), Positives = 16/53 (30%), Gaps = 17/53 (32%)
Frame = -3
Query: 19 PSPPPS-----------------PPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P PPP PP PPPPP P P P P P
Sbjct: 270 PPPPPPLPPLPXSLSPLKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPP 112
>TC226145 weakly similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein
kinase-like protein (CBL-interacting protein kinase 5),
partial (15%)
Length = 890
Score = 40.8 bits (94), Expect = 8e-04
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Frame = -1
Query: 2 PHRV-VLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 55
PHR + +R T P PPP+PP PPP SP+ S SP +SP P+
Sbjct: 509 PHRT*HTPTYLRQT**TPPPPPPAPPRKPPPSSPASPSSPPPSPASSPQTPSSPS 345
Score = 29.3 bits (64), Expect = 2.4
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = -1
Query: 18 SPSPPPSPPVPPPPPSPS 35
SPSP S PPPPPSP+
Sbjct: 353 SPSP*SSASSPPPPPSPT 300
Score = 28.9 bits (63), Expect = 3.1
Identities = 15/41 (36%), Positives = 18/41 (43%)
Frame = -1
Query: 10 PVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 50
P ++ SPPPSP P PS S SP P+P
Sbjct: 425 PPPSSPASPSSPPPSPASSPQTPSSPSP*SSASSPPPPPSP 303
Score = 27.3 bits (59), Expect = 9.0
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 1/47 (2%)
Frame = -1
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLER-SPENSPAPEQQP 54
+P R SP+ P SPP P P SP S S +SP P P
Sbjct: 440 APPRKPPPSSPASPSSPP-PSPASSPQTPSSPSP*SSASSPPPPPSP 303
>TC205429 weakly similar to UP|O22600 (O22600) Glycine-rich protein, partial
(49%)
Length = 611
Score = 40.4 bits (93), Expect = 0.001
Identities = 19/41 (46%), Positives = 21/41 (50%)
Frame = +3
Query: 14 TGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
T V +P PP PPVPP PP P V P SP P+ P
Sbjct: 327 TPVPTPPPPTPPPVPPNPPIPPVVPPRPPRPPPSPPPKPPP 449
Score = 34.7 bits (78), Expect = 0.057
Identities = 16/38 (42%), Positives = 20/38 (52%)
Frame = +3
Query: 17 QSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
++P PPP+P PPPP+P V P N P P P
Sbjct: 306 RTPLPPPTPVPTPPPPTPPPV------PPNPPIPPVVP 401
Score = 33.9 bits (76), Expect = 0.097
Identities = 23/55 (41%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Frame = +3
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVTQEQW--RNLMRSIGNI 71
PSPPP PP P P P+P + L PE +P P P + + W R IGNI
Sbjct: 423 PSPPPKPP-PIPAPTPPKGSPLLPIPE*APTP---P*LMKFFWSLRPASYGIGNI 575
Score = 33.1 bits (74), Expect = 0.16
Identities = 15/41 (36%), Positives = 18/41 (43%)
Frame = +3
Query: 14 TGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
T + P+P P+PP P PPP P P P P P
Sbjct: 309 TPLPPPTPVPTPPPPTPPPVPPNPPIPPVVPPRPPRPPPSP 431
Score = 31.6 bits (70), Expect = 0.48
Identities = 15/39 (38%), Positives = 17/39 (43%)
Frame = +3
Query: 16 VQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
V P PP PP PPP P P + + P PE P
Sbjct: 393 VVPPRPPRPPPSPPPKPPPIPAPTPPKGSPLLPIPE*AP 509
Score = 30.0 bits (66), Expect = 1.4
Identities = 20/56 (35%), Positives = 21/56 (36%)
Frame = +3
Query: 2 PHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
P V P T P+PP P VPP PP P P P P PA T
Sbjct: 321 PPTPVPTPPPPTPPPVPPNPPIPPVVPPRPPRP--------PPSPPPKPPPIPAPT 464
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 40.4 bits (93), Expect = 0.001
Identities = 29/96 (30%), Positives = 38/96 (39%), Gaps = 4/96 (4%)
Frame = +2
Query: 132 DPKDHLLYFN----TKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP HL F+ T D + + FP + + A W L SI N+ D
Sbjct: 575 DPHKHLKEFHIVCSTMKPPDVQEDHIFLKAFPHSLEGVAKDWLYYLAPRSIFNWDDLKRV 754
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARY 223
FL +F D+ IRQ ESL EY R+
Sbjct: 755 FLEKFFPASRTTTIRKDISGIRQLSRESLYEYWERF 862
>BI426606
Length = 357
Score = 40.4 bits (93), Expect = 0.001
Identities = 17/37 (45%), Positives = 21/37 (55%)
Frame = +2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 55
P PPP+PP PPP SP+ S SP +SP P+
Sbjct: 44 PPPPPAPPRKPPPSSPASPSSPPPSPASSPQTPSSPS 154
Score = 28.9 bits (63), Expect = 3.1
Identities = 15/41 (36%), Positives = 18/41 (43%)
Frame = +2
Query: 10 PVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 50
P ++ SPPPSP P PS S SP P+P
Sbjct: 74 PPPSSPASPSSPPPSPASSPQTPSSPSP*SSASSPPPPPSP 196
Score = 27.3 bits (59), Expect = 9.0
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 1/47 (2%)
Frame = +2
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLER-SPENSPAPEQQP 54
+P R SP+ P SPP P P SP S S +SP P P
Sbjct: 59 APPRKPPPSSPASPSSPP-PSPASSPQTPSSPSP*SSASSPPPPPSP 196
>TC228455 similar to UP|Q9M7Y4 (Q9M7Y4) F3E22.8 protein, partial (17%)
Length = 755
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/34 (47%), Positives = 20/34 (58%)
Frame = -2
Query: 21 PPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
PPP PP PPPPP S +R+P + +PE P
Sbjct: 388 PPPLPPNPPPPPKGPSPPSPKRNPSSPASPESSP 287
Score = 36.2 bits (82), Expect = 0.019
Identities = 18/35 (51%), Positives = 21/35 (59%), Gaps = 3/35 (8%)
Frame = -2
Query: 17 QSPSPPPSPPVP---PPPPSPSQVGSLERSPENSP 48
Q P PP+PP P P PPSP + S SPE+SP
Sbjct: 391 QPPPLPPNPPPPPKGPSPPSPKRNPSSPASPESSP 287
Score = 30.4 bits (67), Expect = 1.1
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = -2
Query: 19 PSPPPSPPVPPPPPSPSQVGSLERSPEN 46
P PP P P P +PS S E SP+N
Sbjct: 364 PPPPKGPSPPSPKRNPSSPASPESSPDN 281
>TC204695 similar to UP|Q9LE95 (Q9LE95) Ribosomal protein L1, partial (19%)
Length = 477
Score = 39.7 bits (91), Expect = 0.002
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = +2
Query: 12 RTTGVQSPSPPPSPPVPPPPPS 33
RTT + +P PPP PP PPPPPS
Sbjct: 218 RTTVLMAPPPPPPPPPPPPPPS 283
Score = 34.7 bits (78), Expect = 0.057
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +2
Query: 20 SPPPSPPVPPPPPSPS 35
+PPP PP PPPPP PS
Sbjct: 236 APPPPPPPPPPPPPPS 283
Score = 27.3 bits (59), Expect = 9.0
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +2
Query: 24 SPPVPPPPPSPSQVGSLERS 43
+PP PPPPP P SL ++
Sbjct: 236 APPPPPPPPPPPPPPSLRKA 295
>TC204727 weakly similar to UP|O44358 (O44358) Flagelliform silk protein
(Fragment), partial (5%)
Length = 726
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/45 (44%), Positives = 24/45 (52%)
Frame = -1
Query: 10 PVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P G SP PP PP PP PPSP L +PE +P P+ +P
Sbjct: 330 PSPLPGPPSPPSPPGPPNPPSPPSPEGPPILPSNPEPTP-PKGRP 199
Score = 37.4 bits (85), Expect = 0.009
Identities = 21/51 (41%), Positives = 23/51 (44%)
Frame = -1
Query: 15 GVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVTQEQWRNLM 65
G PSP P PP PP PP P S SPE P P T + R L+
Sbjct: 342 GRLGPSPLPGPPSPPSPPGPPNPPS-PPSPEGPPILPSNPEPTPPKGRPLL 193
>AI794763
Length = 420
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = +3
Query: 11 VRTTGVQSPSPPPSPPVPPPPPSPSQVGSLE-RSPENSPAPEQQPAVTQEQ 60
+R Q PPP PP+P PPP P V ++E P +P P+ V Q Q
Sbjct: 3 LRVRPCQPLRPPPPPPLPSPPPPPPSVINIECPPPPKTPCPDNCEEVPQGQ 155
>TC220926 similar to GB|AAF75234.1|8453100|AF233752 short-root protein
{Arabidopsis thaliana;} , partial (33%)
Length = 937
Score = 39.3 bits (90), Expect = 0.002
Identities = 18/55 (32%), Positives = 27/55 (48%)
Frame = -1
Query: 20 SPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVTQEQWRNLMRSIGNIQQR 74
S PP PP P PP +P+ S + P P+ +P T ++L+R+ QR
Sbjct: 388 STPPGPPPPSPPRAPASASRCSSSQNSHPKPQSKP*TTSNFLQSLLRTRNRRFQR 224
>TC209064 similar to UP|P93166 (P93166) SCOF-1, partial (93%)
Length = 945
Score = 39.3 bits (90), Expect = 0.002
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = +1
Query: 20 SPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 55
+PPP PP P PP P +VG SP+P +P+
Sbjct: 424 NPPPPPPPPAPPTPPPEVGPTSAPSATSPSPPDRPS 531
>TC211133 weakly similar to UP|Q9BKV7 (Q9BKV7) Ppg3, partial (4%)
Length = 709
Score = 39.3 bits (90), Expect = 0.002
Identities = 20/51 (39%), Positives = 26/51 (50%)
Frame = +1
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVTQE 59
SP + +PSP SPP P P SPS S SP +S AP+ A ++
Sbjct: 181 SPPTSATSSTPSPSSSPPSPSPTSSPSSSSSPSPSPPSSAAPDPAAASARD 333
Score = 32.0 bits (71), Expect = 0.37
Identities = 16/46 (34%), Positives = 20/46 (42%)
Frame = +1
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
+P + + S PSP PP PSP+ S SP SP P
Sbjct: 169 APSSSPPTSATSSTPSPSSSPPSPSPTSSPSSSSSPSPSPPSSAAP 306
Score = 30.4 bits (67), Expect = 1.1
Identities = 19/47 (40%), Positives = 21/47 (44%), Gaps = 9/47 (19%)
Frame = +1
Query: 18 SPSPPPSPPV------PPP---PPSPSQVGSLERSPENSPAPEQQPA 55
SP+P SPP P P PPSPS S S SP+P A
Sbjct: 163 SPAPSSSPPTSATSSTPSPSSSPPSPSPTSSPSSSSSPSPSPPSSAA 303
>TC232895 weakly similar to PRF|1604369A.0|226743|1604369A sulfated surface
glycoprotein SSG185. {Volvox carteri;} , partial (14%)
Length = 907
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/55 (40%), Positives = 27/55 (49%), Gaps = 1/55 (1%)
Frame = +3
Query: 2 PHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLE-RSPENSPAPEQQPA 55
P RV + R SPS PP PPPPPSPS L SP + +P +P+
Sbjct: 201 PQRVTSATVSRHAPPTSPSSPPRRNTPPPPPSPSSSTPLRPSSPPPTTSPTSRPS 365
Score = 34.7 bits (78), Expect = 0.057
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Frame = +3
Query: 10 PVRTTGVQSPSPPPSPPVPP--PPPSPSQVGSLERSPENSPAPEQQPA 55
P R T PSP S P+ P PPP+ S R +SP+P PA
Sbjct: 264 PRRNTPPPPPSPSSSTPLRPSSPPPTTSPTSRPSRPTPSSPSPSTAPA 407
Score = 28.9 bits (63), Expect = 3.1
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 4/59 (6%)
Frame = +3
Query: 1 RPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPE----NSPAPEQQPA 55
RP R SP +T +P P+P PP+PS+ + SP P+P +P+
Sbjct: 357 RPSRPTPSSPSPST-----APAPAPTTSTTPPTPSKSKAKPTSPSPTTPTRPSPRVRPS 518
Score = 28.5 bits (62), Expect = 4.1
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 5/54 (9%)
Frame = +3
Query: 9 SPVRTTGVQSPSPPP-----SPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
SP +T ++ SPPP S P P P SP SP +PAP + T
Sbjct: 294 SPSSSTPLRPSSPPPTTSPTSRPSRPTPSSP--------SPSTAPAPAPTTSTT 431
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/40 (42%), Positives = 20/40 (49%)
Frame = +3
Query: 17 QSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAV 56
+ PSPPP PP PPPP PS SP+ P+V
Sbjct: 471 EPPSPPPYPPTSPPPPCPSLKSPNSSPNAASPSKNSSPSV 590
Score = 36.2 bits (82), Expect = 0.019
Identities = 22/56 (39%), Positives = 24/56 (42%), Gaps = 7/56 (12%)
Frame = +3
Query: 9 SPVRTTGVQSPSPPPSP-------PVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
SP T S P PSP P P +PS S R P +SPAP PA T
Sbjct: 222 SPTAVTPPSSSPPRPSPGPSATPTSTSPSPATPSTSSSAPRPPLSSPAPTPSPAPT 389
Score = 32.3 bits (72), Expect = 0.28
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Frame = +3
Query: 16 VQSPSPPPSPP---VPPPPPSPSQVGSLERSPENSPAPEQQP 54
+ SP+P PSP PPPP + S + SP +S A +P
Sbjct: 351 LSSPAPTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEP 476
Score = 28.1 bits (61), Expect = 5.3
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = +3
Query: 13 TTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSP 48
++ + + PPSPP P PP SP +SP +SP
Sbjct: 447 SSSAAATAEPPSPP-PYPPTSPPPPCPSLKSPNSSP 551
Score = 27.7 bits (60), Expect = 6.9
Identities = 16/51 (31%), Positives = 22/51 (42%)
Frame = +3
Query: 4 RVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
R ++ S R+ G S + SP PPPPP + + SP P P
Sbjct: 108 RTLVRSSARS*GTPSQASR-SPAPPPPPPRSASSSTTASSPTAVTPPSSSP 257
>TC234851 weakly similar to UP|Q9DVW0 (Q9DVW0) PxORF73 peptide, partial (30%)
Length = 466
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/49 (40%), Positives = 22/49 (44%)
Frame = +3
Query: 9 SPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
SP + SPSPP S P PPP P + SL P P P A T
Sbjct: 288 SPSSASRPSSPSPPTSSPSPPPYPPSTPPSSLSTPPPPPPPPPPPSATT 434
Score = 33.5 bits (75), Expect = 0.13
Identities = 18/41 (43%), Positives = 18/41 (43%), Gaps = 8/41 (19%)
Frame = +3
Query: 18 SPSPPPSPP--------VPPPPPSPSQVGSLERSPENSPAP 50
SPSPPP PP PPPPP P S SP P
Sbjct: 336 SPSPPPYPPSTPPSSLSTPPPPPPPPPPPSATTPRAFSPPP 458
Score = 32.0 bits (71), Expect = 0.37
Identities = 17/34 (50%), Positives = 17/34 (50%), Gaps = 5/34 (14%)
Frame = +3
Query: 19 PSPPPS-----PPVPPPPPSPSQVGSLERSPENS 47
PS PPS PP PPPPP PS SP S
Sbjct: 360 PSTPPSSLSTPPPPPPPPPPPSATTPRAFSPPPS 461
Score = 32.0 bits (71), Expect = 0.37
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = +3
Query: 13 TTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPEN 46
+T S S PP PP PPPPPS + + P +
Sbjct: 363 STPPSSLSTPPPPPPPPPPPSATTPRAFSPPPSS 464
Score = 27.3 bits (59), Expect = 9.0
Identities = 15/37 (40%), Positives = 17/37 (45%)
Frame = +3
Query: 21 PPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
P PSPP P PP S S R SP +PA +
Sbjct: 168 PNPSPPTPSPPKS-----STWRPSHTSPPHVSRPATS 263
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/48 (39%), Positives = 25/48 (51%), Gaps = 3/48 (6%)
Frame = +1
Query: 10 PVRTTGVQSPSP---PPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P T ++PSP PP+ P PP PSPS +P +P+P QP
Sbjct: 64 PYVPTPPKTPSPSYSPPNVPTPPKTPSPSSQPPFTPTPPKTPSPTSQP 207
Score = 35.8 bits (81), Expect = 0.025
Identities = 18/43 (41%), Positives = 22/43 (50%), Gaps = 3/43 (6%)
Frame = +1
Query: 18 SPSP---PPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
+PSP PP P PP PSPS +P +P+P QP T
Sbjct: 40 APSPGNHPPYVPTPPKTPSPSYSPPNVPTPPKTPSPSSQPPFT 168
Score = 35.8 bits (81), Expect = 0.025
Identities = 17/45 (37%), Positives = 22/45 (48%)
Frame = +1
Query: 10 PVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
P T ++PSP PP P PPSP +P N+ +P QP
Sbjct: 160 PFTPTPPKTPSPTSQPPYIPTPPSPISQPPSIATPPNTLSPTSQP 294
Score = 33.1 bits (74), Expect = 0.16
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 7/50 (14%)
Frame = +1
Query: 17 QSPSPPPSPPVPPPP-------PSPSQVGSLERSPENSPAPEQQPAVTQE 59
++PSP +PP+ P P P+P + S SP N P P + P+ + +
Sbjct: 7 KTPSPIYAPPIAPSPGNHPPYVPTPPKTPSPSYSPPNVPTPPKTPSPSSQ 156
Score = 28.1 bits (61), Expect = 5.3
Identities = 14/41 (34%), Positives = 17/41 (41%)
Frame = +1
Query: 14 TGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 54
T ++PSP PP P PP S P+P QP
Sbjct: 124 TPPKTPSPSSQPPFTPTPPKTPSPTSQPPYIPTPPSPISQP 246
>TC228334 similar to UP|Q9SW80 (Q9SW80) Bel1-like homeodomain 2
(AT4g36870/C7A10_490), partial (9%)
Length = 910
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = +2
Query: 6 VLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAVT 57
++D R T +SP PP PP PPPPP + L S + P + P +T
Sbjct: 272 IIDKASRKTKQRSPPHPPPPPPPPPPPLRWRHPCLSASTQTCPHTD*WPLMT 427
>TC226338 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (92%)
Length = 860
Score = 38.9 bits (89), Expect = 0.003
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 5/67 (7%)
Frame = +2
Query: 7 LDSPVRTT-GVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSP----APEQQPAVTQEQW 61
L P +TT ++SP+P +P PPPPP P L+ S + P AP+ P+++
Sbjct: 131 LRRPRKTTLSLRSPNPSSTPAPPPPPPPPRFPNLLDSSMQPPPPLHRAPQSDPSLSTRNL 310
Query: 62 RNLMRSI 68
R R +
Sbjct: 311 RRTKRRL 331
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.131 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,866,736
Number of Sequences: 63676
Number of extensions: 278648
Number of successful extensions: 26025
Number of sequences better than 10.0: 1476
Number of HSP's better than 10.0 without gapping: 6780
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 15111
length of query: 310
length of database: 12,639,632
effective HSP length: 97
effective length of query: 213
effective length of database: 6,463,060
effective search space: 1376631780
effective search space used: 1376631780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0371b.8


