

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.12
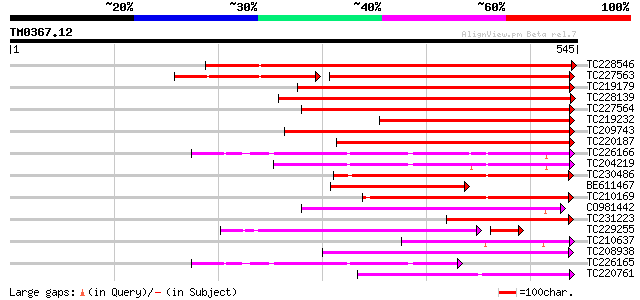
(545 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228546 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partia... 633 0.0
TC227563 similar to PIR|E96542|E96542 scarecrow-like protein [im... 309 e-127
TC219179 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 394 e-110
TC228139 similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible gibberel... 363 e-101
TC227564 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 345 4e-95
TC219232 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 304 6e-83
TC209743 similar to PIR|E86347|E86347 scarecrow-like 1 protein F... 302 3e-82
TC220187 similar to PIR|E86347|E86347 scarecrow-like 1 protein F... 243 1e-64
TC226166 similar to UP|O23724 (O23724) GAI protein, partial (70%) 203 2e-52
TC204219 similar to UP|Q7Y1B6 (Q7Y1B6) GAI-like protein, partial... 202 3e-52
TC230486 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional... 181 7e-46
BE611467 176 3e-44
TC210169 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional... 169 4e-42
CO981442 161 6e-40
TC231223 weakly similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible g... 154 1e-37
TC229255 similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like protein, p... 130 3e-37
TC210637 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional... 142 3e-34
TC208938 weakly similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like pro... 141 8e-34
TC226165 GAI1 [Glycine max] 131 9e-31
TC220761 similar to UP|Q7G7J6 (Q7G7J6) Gibberellin-insensitive p... 126 2e-29
>TC228546 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial (60%)
Length = 1493
Score = 633 bits (1633), Expect = 0.0
Identities = 309/357 (86%), Positives = 332/357 (92%)
Frame = +3
Query: 189 TALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSLIYKALKCEQPITSKELMS 248
TA+G+MNNVL KMVSV GDPIQRLGAYMLEGLRARLESSGS+IYKALKCEQP TS +LM+
Sbjct: 3 TAVGFMNNVLAKMVSVGGDPIQRLGAYMLEGLRARLESSGSIIYKALKCEQP-TSNDLMT 179
Query: 249 YMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGP 308
YM ILYQ+CPYWKF Y S+N VIGEAM NESRI IIDFQI QGTQWLLLIQALASRPGGP
Sbjct: 180 YMHILYQICPYWKFAYTSANAVIGEAMLNESRIRIIDFQIAQGTQWLLLIQALASRPGGP 359
Query: 309 PFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRP 368
PF+ VTGVDDS SF ARGGGLHIVGKRLSD+AKSCGVPFEFHSAAM G E+ELENLVI+P
Sbjct: 360 PFVHVTGVDDSQSFHARGGGLHIVGKRLSDYAKSCGVPFEFHSAAMCGSELELENLVIQP 539
Query: 369 GEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFV 428
GEAL VNF F LHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFV
Sbjct: 540 GEALVVNFPFVLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFV 719
Query: 429 ETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSR 488
ETLSYY+AM+ESIDVALPRDDK RI+AEQHCVARDIVNM+ACEG ER+ERHEL GKWRSR
Sbjct: 720 ETLSYYTAMFESIDVALPRDDKQRINAEQHCVARDIVNMVACEGDERLERHELLGKWRSR 899
Query: 489 FSMAGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVALYLTWKNRAMCTASAWRCF 545
FSMAGF PCPLSSSVTA+VRN+LNEFNENYRL+H+D ALYL WK+RAMCT+SAWRC+
Sbjct: 900 FSMAGFAPCPLSSSVTAAVRNMLNEFNENYRLQHRDGALYLGWKSRAMCTSSAWRCY 1070
>TC227563 similar to PIR|E96542|E96542 scarecrow-like protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(71%)
Length = 1274
Score = 309 bits (792), Expect(2) = e-127
Identities = 149/236 (63%), Positives = 184/236 (77%)
Frame = +2
Query: 308 PPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIR 367
PP IR+TG DDS S AR GGL IVG RLS A+S VPFEFH+ + EVEL++L ++
Sbjct: 449 PPKIRITGFDDSTSAYAREGGLEIVGARLSRLAQSYNVPFEFHAIRAAPTEVELKDLALQ 628
Query: 368 PGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRF 427
PGEA+AVNFA LHH+PDE V + NHRDRL+RL K LSPK+VTLVEQES+TN PFF RF
Sbjct: 629 PGEAIAVNFAMMLHHVPDECVDSRNHRDRLVRLAKCLSPKIVTLVEQESHTNNLPFFPRF 808
Query: 428 VETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRS 487
VET++YY A++ESIDVALPR+ K RI+ EQHC+AR++VN+IACEGAERVERHEL KWRS
Sbjct: 809 VETMNYYLAIFESIDVALPREHKERINVEQHCLAREVVNLIACEGAERVERHELLKKWRS 988
Query: 488 RFSMAGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVALYLTWKNRAMCTASAWR 543
RF+MAGF P PL+S VT S++N+ + +Y LE +D AL L W N+ + T+ AWR
Sbjct: 989 RFTMAGFTPYPLNSFVTCSIKNLQQSYQGHYTLEERDGALCLGWMNQVLITSCAWR 1156
Score = 164 bits (414), Expect(2) = e-127
Identities = 82/140 (58%), Positives = 106/140 (75%)
Frame = +3
Query: 159 WAQIEEMIPKLDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLE 218
W ++ EMI + DLK++L CA+AV+ DM+T W+ + L KMVSV+G+PIQRLGAYMLE
Sbjct: 6 WKRMMEMISRGDLKEMLCTCAKAVAGNDMETT-EWLMSELRKMVSVSGNPIQRLGAYMLE 182
Query: 219 GLRARLESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNE 278
L ARL SSGS IYK LKC++P T EL+S+M +LY++CPY KF Y+S+N I E M+ E
Sbjct: 183 ALVARLASSGSTIYKVLKCKEP-TGSELLSHMHLLYEICPYLKFGYMSANGAIAEVMKEE 359
Query: 279 SRIHIIDFQITQGTQWLLLI 298
S +HIIDFQI QG QW+ LI
Sbjct: 360 SEVHIIDFQINQGIQWVSLI 419
>TC219179 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(52%)
Length = 1025
Score = 394 bits (1012), Expect = e-110
Identities = 186/267 (69%), Positives = 225/267 (83%)
Frame = +3
Query: 277 NESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRL 336
+E R+HIIDFQI QG+QW+ LIQA A+RPGGPP IR+TG+DDS S ARGGGLHIVG+RL
Sbjct: 3 DEDRVHIIDFQIGQGSQWITLIQAFAARPGGPPHIRITGIDDSTSAYARGGGLHIVGRRL 182
Query: 337 SDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDR 396
S A+ VPFEFH+AA+SG +V+L NL +RPGEALAVNFAF LHHMPDESVST+NHRDR
Sbjct: 183 SKLAEHFKVPFEFHAAAISGFDVQLHNLGVRPGEALAVNFAFMLHHMPDESVSTQNHRDR 362
Query: 397 LLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAE 456
LLRLV+SLSPKVVTLVEQESNTNT+ FF RF+ETL+YY+AM+ESIDV LPR+ K RI+ E
Sbjct: 363 LLRLVRSLSPKVVTLVEQESNTNTAAFFPRFLETLNYYTAMFESIDVTLPREHKERINVE 542
Query: 457 QHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNE 516
QHC+ARD+VN+IACEG ERVERHE+ GKWRSRF+MAGF P PLSS V +++ +L +++
Sbjct: 543 QHCLARDLVNIIACEGVERVERHEVLGKWRSRFAMAGFTPYPLSSLVNGTIKKLLENYSD 722
Query: 517 NYRLEHKDVALYLTWKNRAMCTASAWR 543
YRLE +D ALYL W NR + + AW+
Sbjct: 723 RYRLEERDGALYLGWMNRDLVASCAWK 803
>TC228139 similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible
gibberellin-responsive protein, partial (47%)
Length = 1497
Score = 363 bits (933), Expect = e-101
Identities = 178/286 (62%), Positives = 216/286 (75%)
Frame = +1
Query: 259 YWKFTYISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDD 318
Y KF Y+++N I +A +NE IHIIDFQI QGTQW+ L+QALA+RPGG P +R+TG+DD
Sbjct: 4 YLKFGYMAANGAIAQACRNEDHIHIIDFQIAQGTQWMTLLQALAARPGGAPHVRITGIDD 183
Query: 319 SLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAF 378
+S ARG GL +VGKRL+ ++ G+P EFH + V E L IRPGEALAVNF
Sbjct: 184 PVSKYARGDGLEVVGKRLALMSEKFGIPVEFHGVPVFAPNVTREMLDIRPGEALAVNFPL 363
Query: 379 FLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMY 438
LHH DESV N RD LLRLV+SLSPKV TLVEQESNTNT+PFF RF+ETL YY A++
Sbjct: 364 QLHHTADESVHVSNPRDGLLRLVRSLSPKVTTLVEQESNTNTTPFFNRFIETLDYYLAIF 543
Query: 439 ESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCP 498
ESIDV LPRD K RI+ EQHC+ARDIVN+IACEG ERVERHELFGKW+SR MAGF CP
Sbjct: 544 ESIDVTLPRDSKERINVEQHCLARDIVNIIACEGKERVERHELFGKWKSRLKMAGFQQCP 723
Query: 499 LSSSVTASVRNILNEFNENYRLEHKDVALYLTWKNRAMCTASAWRC 544
LSS V + +R++L ++E+Y L KD A+ L WK+R + +ASAW C
Sbjct: 724 LSSYVNSVIRSLLMCYSEHYTLVEKDGAMLLGWKDRNLISASAWHC 861
>TC227564 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(53%)
Length = 1107
Score = 345 bits (884), Expect = 4e-95
Identities = 165/263 (62%), Positives = 208/263 (78%)
Frame = +3
Query: 281 IHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFA 340
+HI+DFQI QGTQW+ LIQALA RP GPP IR++GVDDS S AR GGL IVGKRLS A
Sbjct: 6 VHIVDFQIGQGTQWVSLIQALARRPVGPPKIRISGVDDSYSAYARRGGLDIVGKRLSALA 185
Query: 341 KSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRL 400
+SC VPFEF++ + EV+LE+L +RP EA+AVNFA LHH+PDESV++ NHRDRLLRL
Sbjct: 186 QSCHVPFEFNAVRVPVTEVQLEDLELRPYEAVAVNFAISLHHVPDESVNSHNHRDRLLRL 365
Query: 401 VKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCV 460
K LSPKVVTLVEQE +TN +PF QRFVET++YY A++ESID LPR+ K RI+ EQHC+
Sbjct: 366 AKQLSPKVVTLVEQEFSTNNAPFLQRFVETMNYYLAVFESIDTVLPREHKERINVEQHCL 545
Query: 461 ARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYRL 520
AR++VN+IACEG ERVERHEL KWR RF+ AGF P PLSS + +S++++L ++ +Y L
Sbjct: 546 AREVVNLIACEGEERVERHELLNKWRMRFTKAGFTPYPLSSVINSSIKDLLQSYHGHYTL 725
Query: 521 EHKDVALYLTWKNRAMCTASAWR 543
E +D AL+L W N+ + + AWR
Sbjct: 726 EERDGALFLGWMNQVLVASCAWR 794
>TC219232 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(38%)
Length = 881
Score = 304 bits (779), Expect = 6e-83
Identities = 147/188 (78%), Positives = 162/188 (85%)
Frame = +3
Query: 356 GCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQE 415
GCEV N+ + PGEALAV+F + LHHMPDESVSTENHRDRLLRLVK LSPKVVT+VEQE
Sbjct: 3 GCEVVRGNIEVLPGEALAVSFPYVLHHMPDESVSTENHRDRLLRLVKRLSPKVVTIVEQE 182
Query: 416 SNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAER 475
SNTNTSPFF RFVETL YY+AM+ESIDVA PRDDK RISAEQHCVARDIVNMIACEG ER
Sbjct: 183 SNTNTSPFFHRFVETLDYYTAMFESIDVACPRDDKKRISAEQHCVARDIVNMIACEGVER 362
Query: 476 VERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVALYLTWKNRA 535
VERHEL GKWRSR SMAGF C LSSSV +++N+L EF++NYRLEH+D ALYL W NR
Sbjct: 363 VERHELLGKWRSRLSMAGFKQCQLSSSVMVAIQNLLKEFSQNYRLEHRDGALYLGWMNRH 542
Query: 536 MCTASAWR 543
M T+SAWR
Sbjct: 543 MATSSAWR 566
>TC209743 similar to PIR|E86347|E86347 scarecrow-like 1 protein F24J8.8 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(47%)
Length = 1078
Score = 302 bits (773), Expect = 3e-82
Identities = 140/280 (50%), Positives = 206/280 (73%), Gaps = 1/280 (0%)
Frame = +1
Query: 265 ISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDA 324
+ +N I E +++E ++HIIDF I+QGTQ++ LIQ LAS PG PP +R+T VDD S
Sbjct: 7 VGANGAIAEVVRDEKKVHIIDFDISQGTQYITLIQTLASMPGRPPHVRLTAVDDPESVQR 186
Query: 325 RGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMP 384
GG++I+G+RL A+ +PFEF + A V L RPGEAL VNFAF LHHM
Sbjct: 187 SIGGINIIGQRLEKLAEELRLPFEFRAVASRTSNVTQSMLNCRPGEALVVNFAFQLHHMR 366
Query: 385 DESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVA 444
DE+VST N RD+LLR+VKSL+PK+VT+VEQ+ NTNTSPF RF+ET +YYSA++ ++D
Sbjct: 367 DETVSTVNERDQLLRMVKSLNPKIVTVVEQDMNTNTSPFLPRFIETYNYYSAVFNTLDAT 546
Query: 445 LPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVT 504
LPR+ ++R++ E+ C+A+DIVN++ACEG ER+ER+E+ GKWR+R SMAGF P P+S++V
Sbjct: 547 LPRESQDRMNVERQCLAKDIVNIVACEGEERIERYEVAGKWRARLSMAGFTPSPMSTNVR 726
Query: 505 ASVRN-ILNEFNENYRLEHKDVALYLTWKNRAMCTASAWR 543
++RN I+ ++ + ++++ + L+ W+++ + ASAW+
Sbjct: 727 EAIRNLIIKQYCDKFKIKEEMGGLHFGWEDKNLIVASAWK 846
>TC220187 similar to PIR|E86347|E86347 scarecrow-like 1 protein F24J8.8 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(39%)
Length = 810
Score = 243 bits (621), Expect = 1e-64
Identities = 117/230 (50%), Positives = 166/230 (71%), Gaps = 1/230 (0%)
Frame = +3
Query: 315 GVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAV 374
GVDD S GGL +G+RL A++ G+PFEF + A V L P EAL V
Sbjct: 3 GVDDPESVQRSVGGLQNIGQRLEKLAEALGLPFEFRAVASRTSIVTPSMLNCSPDEALVV 182
Query: 375 NFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYY 434
NFAF LHHMPDESVST N RD+LLRLVKSL+PK+VT+VEQ+ NTNT+PF RFVE +YY
Sbjct: 183 NFAFQLHHMPDESVSTANERDQLLRLVKSLNPKLVTVVEQDVNTNTTPFLPRFVEAYNYY 362
Query: 435 SAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGF 494
SA++ES+D LPR+ ++R++ E+ C+ARDIVN++ACEG +R+ER+E+ GKWR+R +MAGF
Sbjct: 363 SAVFESLDATLPRESQDRMNVERQCLARDIVNVVACEGEDRIERYEVAGKWRARMTMAGF 542
Query: 495 VPCPLSSSVTASVRNILNE-FNENYRLEHKDVALYLTWKNRAMCTASAWR 543
P+S++VT +R ++ + + Y+++ + AL+ W+++ + ASAW+
Sbjct: 543 TSSPMSTNVTDEIRKLIKTVYCDRYKIKEEMGALHFGWEDKNLIVASAWK 692
>TC226166 similar to UP|O23724 (O23724) GAI protein, partial (70%)
Length = 1613
Score = 203 bits (516), Expect = 2e-52
Identities = 121/372 (32%), Positives = 202/372 (53%), Gaps = 3/372 (0%)
Frame = +1
Query: 175 LIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSLIYKA 234
L+ CA+AV + ++ A + + VS G ++++ Y E L R IY+
Sbjct: 124 LMACAEAVENNNLAVAEALVKQIGFLAVSQVG-AMRKVAIYFAEALARR-------IYRV 279
Query: 235 LKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQITQGTQW 294
+ ++ + + Y+ CPY KF + ++N VI EA Q ++R+H+IDF I QG QW
Sbjct: 280 FPLQHSLSDSLQIHF----YETCPYLKFAHFTANQVILEAFQGKNRVHVIDFGINQGMQW 447
Query: 295 LLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHS-AA 353
L+QALA R GGPP R+TG+ + ++ L VG +L+ A+ V FE+ A
Sbjct: 448 PALMQALAVRTGGPPVFRLTGIGPPAADNS--DHLQEVGWKLAQLAEEINVQFEYRGFVA 621
Query: 354 MSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVE 413
S +++ L +R E++AVN F H + ++ +++L +V+ + P+++T+VE
Sbjct: 622 NSLADLDASMLDLREDESVAVNSVFEFHKL----LARPGAVEKVLSVVRQIRPEILTVVE 789
Query: 414 QESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGA 473
QE+N N F RF E+L YYS +++S++ P + ++ +E + + + I N++ACEG
Sbjct: 790 QEANHNGLSFVDRFTESLHYYSTLFDSLE-GSPVNPNDKAMSEVY-LGKQICNVVACEGM 963
Query: 474 ERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEF--NENYRLEHKDVALYLTW 531
+RVERHE +WR+RF GF P L S+ +L+ F + YR+E + L L W
Sbjct: 964 DRVERHETLNQWRNRFGSTGFSPVHLGSNAYKQASMLLSLFGGGDGYRVEENNGCLMLGW 1143
Query: 532 KNRAMCTASAWR 543
R + S W+
Sbjct: 1144HTRPLIATSVWQ 1179
>TC204219 similar to UP|Q7Y1B6 (Q7Y1B6) GAI-like protein, partial (50%)
Length = 1208
Score = 202 bits (514), Expect = 3e-52
Identities = 116/298 (38%), Positives = 169/298 (55%), Gaps = 8/298 (2%)
Frame = +1
Query: 254 YQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRV 313
Y+ CPY KF + ++N I EA ++H+IDF + QG QW L+QALA RPGGPP R+
Sbjct: 19 YESCPYLKFAHFTANQAILEAFATAGKVHVIDFGLKQGMQWPALMQALALRPGGPPTFRL 198
Query: 314 TGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHS-AAMSGCEVELENLVIRPGEAL 372
TG+ + L VG +L+ A++ GV FEF S +++ + L IRPGEA+
Sbjct: 199 TGIGPPQPDNT--DALQQVGWKLAQLAQNIGVQFEFRGFVCNSLADLDPKMLEIRPGEAV 372
Query: 373 AVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLS 432
AVN F LH M ++ D++L VK + PK+VT+VEQE+N N F RF E L
Sbjct: 373 AVNSVFELHRM----LARPGSVDKVLDTVKKIKPKIVTIVEQEANHNGPGFLDRFTEALH 540
Query: 433 YYSAMYESID-----VALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRS 487
YYS++++S++ L +++ + +E + + R I N++A EGA+RVERHE +WR
Sbjct: 541 YYSSLFDSLEGSSSSTGLGSPNQDLLMSELY-LGRQICNVVANEGADRVERHETLSQWRG 717
Query: 488 RFSMAGFVPCPLSSSVTASVRNILNEF--NENYRLEHKDVALYLTWKNRAMCTASAWR 543
R AGF P L S+ +L F + YR+E + L L W R + SAW+
Sbjct: 718 RLDSAGFDPVHLGSNAFKQASMLLALFAGGDGYRVEENNGCLMLGWHTRPLIATSAWK 891
>TC230486 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional
regulator-like, partial (44%)
Length = 886
Score = 181 bits (459), Expect = 7e-46
Identities = 99/231 (42%), Positives = 144/231 (61%)
Frame = +2
Query: 312 RVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEA 371
++ V ++ + D R L+IVG L A+ G+ FEF E+ E+L E
Sbjct: 2 KIVVVTENCADDER---LNIVGVLLGRHAEKLGIGFEFKVLTRRIAELTRESLGCDADEP 172
Query: 372 LAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETL 431
LAVNFA+ L+ MPDESVSTEN RD+LLR VK+L+P+VVTLVEQ++N NT+PF R E
Sbjct: 173 LAVNFAYKLYRMPDESVSTENPRDKLLRRVKTLAPRVVTLVEQDANANTAPFVARVTELC 352
Query: 432 SYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSM 491
+YY A+++S++ + R++ R+ E+ ++R +VN +ACEG +RVER E+FGKWR+R SM
Sbjct: 353 AYYGALFDSLESTMARENLKRVRIEEG-LSRKVVNSVACEGRDRVERCEVFGKWRARMSM 529
Query: 492 AGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVALYLTWKNRAMCTASAW 542
AGF PLS V S++ L ++ ++ + W R + ASAW
Sbjct: 530 AGFRLKPLSQRVADSIKARLGGAGNRVAVKVENGGICFGWMGRTLTVASAW 682
>BE611467
Length = 412
Score = 176 bits (445), Expect = 3e-44
Identities = 89/134 (66%), Positives = 106/134 (78%)
Frame = +3
Query: 309 PFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRP 368
P IR++GVDDS S ARGGGL IVGKRLS A+SC VPFEF++ + +V+LE+L + P
Sbjct: 3 PKIRISGVDDSYSAYARGGGLDIVGKRLSAHAQSCHVPFEFNAVRVPASQVQLEDLELLP 182
Query: 369 GEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFV 428
EA+AVNFA LHH+PDESV++ NHRDRLLRL K LSPKVVTLVEQE NTN +PF QRF
Sbjct: 183 YEAVAVNFAISLHHVPDESVNSHNHRDRLLRLAKRLSPKVVTLVEQEFNTNNAPFLQRFD 362
Query: 429 ETLSYYSAMYESID 442
ET+ YY A+ ESID
Sbjct: 363 ETMKYYLAVXESID 404
>TC210169 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional
regulator-like, partial (47%)
Length = 872
Score = 169 bits (427), Expect = 4e-42
Identities = 92/203 (45%), Positives = 127/203 (62%)
Frame = +1
Query: 340 AKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLR 399
A+ C FEF E+ E+L EALAVNFA+ L+ MPDESVSTEN RD LLR
Sbjct: 79 ARDC---FEFKVLIRRIAELTRESLDCDADEALAVNFAYKLYRMPDESVSTENPRDELLR 249
Query: 400 LVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHC 459
VK+L+P+VVTLVEQE+N NT+PF R E +YY A+++S++ + R++ R+ E+
Sbjct: 250 RVKALAPRVVTLVEQEANANTAPFVARVSELCAYYGALFDSLESTMARENSARVRIEEG- 426
Query: 460 VARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYR 519
++R + N +ACEG RVER E+FGKWR+R SMAGF PLS V S++ L
Sbjct: 427 LSRKVGNSVACEGRNRVERCEVFGKWRARMSMAGFRLKPLSQRVAESIKARLGGAGNRVA 606
Query: 520 LEHKDVALYLTWKNRAMCTASAW 542
++ ++ + W R + ASAW
Sbjct: 607 VKVENGGICFGWMGRTLTVASAW 675
>CO981442
Length = 791
Score = 161 bits (408), Expect = 6e-40
Identities = 93/256 (36%), Positives = 139/256 (53%), Gaps = 2/256 (0%)
Frame = -2
Query: 281 IHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFA 340
IHIIDF I G +W LI L+ R GGPP +R+TG+D + G+RL++F
Sbjct: 769 IHIIDFGIRYGFKWPALISRLSRRSGGPPKLRITGIDVPQPGLRPQERVLETGRRLANFC 590
Query: 341 KSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRL 400
K VPFEF++ A + +E+L I P E +AVN F H+ DE+V N RD +LRL
Sbjct: 589 KRFNVPFEFNAIAQRWDTIRVEDLKIEPNEFVAVNCLFQFEHLLDETVVLNNSRDAVLRL 410
Query: 401 VKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCV 460
+K+ +P + + + F RF E L +Y+A+++ +D + R D R+ E+
Sbjct: 409 IKNANPDIFVHGIVNGSYDVPFFVSRFREALFHYTALFDMLDTNVARQDPMRLMFEKELF 230
Query: 461 ARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNE--FNENY 518
R+IVN+IACEG ERVER + + +W+ R GF PL + +++ L + N N+
Sbjct: 229 GREIVNIIACEGFERVERPQTYKQWQLRNMRNGFRLLPLDHRIIGKLKDRLRDDAHNNNF 50
Query: 519 RLEHKDVALYLTWKNR 534
LE + WK R
Sbjct: 49 LLEVDGDWVLQGWKGR 2
>TC231223 weakly similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible
gibberellin-responsive protein, partial (22%)
Length = 653
Score = 154 bits (388), Expect = 1e-37
Identities = 71/122 (58%), Positives = 95/122 (77%)
Frame = +1
Query: 421 SPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHE 480
+PFF RF+ETL YY A++ESIDV+LPR K +I+ EQHC+ARDIVN+IACEG ERVERHE
Sbjct: 10 TPFFNRFIETLDYYLAIFESIDVSLPRKSKVQINMEQHCLARDIVNIIACEGKERVERHE 189
Query: 481 LFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVALYLTWKNRAMCTAS 540
L GKW+SR +MAGF PLSS + + +R++L ++++Y L KD A+ L WK+R + + S
Sbjct: 190 LLGKWKSRLTMAGFRQYPLSSYMNSVIRSLLRCYSKHYNLVEKDGAMLLGWKDRNLISTS 369
Query: 541 AW 542
AW
Sbjct: 370 AW 375
>TC229255 similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like protein, partial (37%)
Length = 1561
Score = 130 bits (326), Expect(2) = 3e-37
Identities = 76/251 (30%), Positives = 131/251 (51%)
Frame = +1
Query: 203 SVAGDPIQRLGAYMLEGLRARLESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYWKF 262
S GD +QRL Y GL+ RL ++G+ Y L + TS +++ + P +
Sbjct: 64 SAFGDGLQRLAHYFANGLQTRL-AAGTPSYTPL---EGTTSADMLKAYKLYVTSSPLQRL 231
Query: 263 TYISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSF 322
T + I ++NE +HIIDF I G QW LI+ L+ R GGPP +R+TG++
Sbjct: 232 TNYLATKTIVSLVENEGSVHIIDFGICYGFQWPCLIKKLSERHGGPPRLRITGIELPQPG 411
Query: 323 DARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHH 382
+ G+RL+++ K VPFE++ A ++L +L I E V+ + L +
Sbjct: 412 FRPAERVEETGRRLANYCKKFKVPFEYNCLAQKWETIKLADLKIDRNEVTVVSCFYRLKN 591
Query: 383 MPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESID 442
+PDE+V ++ RD +L+L++ ++P + N F RF E L ++S++++ +
Sbjct: 592 LPDETVDVKSPRDAVLKLIRRINPNMFIHGVVNGTYNAPFFLTRFREALYHFSSLFDMFE 771
Query: 443 VALPRDDKNRI 453
+PR+D R+
Sbjct: 772 ANVPREDPERV 804
Score = 43.9 bits (102), Expect(2) = 3e-37
Identities = 19/32 (59%), Positives = 24/32 (74%)
Frame = +3
Query: 463 DIVNMIACEGAERVERHELFGKWRSRFSMAGF 494
D +N+IACEGAERVER E + +W+ R AGF
Sbjct: 804 DAINVIACEGAERVERPETYKQWQVRNQRAGF 899
>TC210637 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional
regulator-like, partial (43%)
Length = 540
Score = 142 bits (359), Expect = 3e-34
Identities = 78/175 (44%), Positives = 105/175 (59%), Gaps = 8/175 (4%)
Frame = +2
Query: 377 AFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSA 436
AF +PDESVSTEN RD L R VK L +VVT+VEQE N NT+PF R LSYYSA
Sbjct: 5 AFXXXKIPDESVSTENPRDELXRRVKRLXXRVVTIVEQEINANTAPFLARVAXXLSYYSA 184
Query: 437 MYESIDVALPRDDKNRISAE----QHCVARDIVNMIACEGAERVERHELFGKWRSRFSMA 492
+ ESI+ + N + + + ++R + N +ACEG +RVER E+FGKWR+R SMA
Sbjct: 185 LLESIEATTAGRENNNNNLDRVRLEEGLSRKLHNSVACEGRDRVERCEVFGKWRARMSMA 364
Query: 493 GFVPCPLSSSVTASVRNIL----NEFNENYRLEHKDVALYLTWKNRAMCTASAWR 543
GF PLS S+ S+++ L N N ++ ++ + W R + ASAWR
Sbjct: 365 GFELKPLSQSMAESIKSRLTTANNRVNSGLTVKEENGGICFGWMGRTLTVASAWR 529
>TC208938 weakly similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like protein,
partial (31%)
Length = 915
Score = 141 bits (355), Expect = 8e-34
Identities = 78/243 (32%), Positives = 129/243 (52%), Gaps = 1/243 (0%)
Frame = +2
Query: 301 LASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVE 360
L+ RPGGPP +R+ G+D + G+ L + K GVPFE++ A +
Sbjct: 2 LSERPGGPPKLRMMGIDLPQPGFRPAERVEETGRWLEKYCKRFGVPFEYNCLAQKWETIR 181
Query: 361 LENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNT 420
LE+L I E VN + L ++ DE+V+ RD LLRL++ ++P + N
Sbjct: 182 LEDLKIDRSEVTVVNCLYRLKNLSDETVTANCPRDALLRLIRRINPNIFMHGVVNGTYNA 361
Query: 421 SPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHE 480
F RF E L ++S++++ + +PR+D +R+ E+ RD +N+IACEGAERVER E
Sbjct: 362 PFFVTRFREALFHFSSLFDMFEANVPREDPSRLMIEKGLFGRDAINVIACEGAERVERPE 541
Query: 481 LFGKWRSRFSMAGFVPCPLSSSVTASVRNIL-NEFNENYRLEHKDVALYLTWKNRAMCTA 539
+ +W+ R AGF PL+ V+ ++ E ++++ ++ + WK R +
Sbjct: 542 TYKQWQVRNQRAGFKQLPLAPEHVNRVKEMVKKEHHKDFVVDEDGKWVLQGWKGRILFAV 721
Query: 540 SAW 542
S+W
Sbjct: 722 SSW 730
>TC226165 GAI1 [Glycine max]
Length = 1313
Score = 131 bits (329), Expect = 9e-31
Identities = 83/262 (31%), Positives = 140/262 (52%), Gaps = 1/262 (0%)
Frame = +3
Query: 175 LIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSLIYKA 234
L+ CA+AV + ++ A + + +S G ++++ Y E L R IY+
Sbjct: 579 LMACAEAVENNNLAVAEALVKQIGFLALSQVG-AMRKVATYFAEALARR-------IYRV 734
Query: 235 LKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQITQGTQW 294
+ ++ + + Y+ CPY KF + ++N I EA Q ++R+H+IDF I QG QW
Sbjct: 735 FPQQHSLSDSLQIHF----YETCPYLKFAHFTANQAILEAFQGKNRVHVIDFGINQGMQW 902
Query: 295 LLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFHS-AA 353
L+QALA R GPP R+TG+ + ++ L VG +L+ A+ V FE+ A
Sbjct: 903 PALMQALALRNDGPPVFRLTGIGPPAADNS--DHLQEVGWKLAQLAERIHVQFEYRGFVA 1076
Query: 354 MSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVE 413
S +++ L +R E++AVN F H + ++ +++L +V+ + P+++T+VE
Sbjct: 1077NSLADLDASMLDLREDESVAVNSVFEFHKL----LARPGAVEKVLSVVRQIRPEILTVVE 1244
Query: 414 QESNTNTSPFFQRFVETLSYYS 435
QE+N N F RF E+L YYS
Sbjct: 1245QEANHNGLSFVDRFTESLHYYS 1310
>TC220761 similar to UP|Q7G7J6 (Q7G7J6) Gibberellin-insensitive protein
OsGAI, partial (23%)
Length = 719
Score = 126 bits (317), Expect = 2e-29
Identities = 76/212 (35%), Positives = 114/212 (52%), Gaps = 3/212 (1%)
Frame = +3
Query: 335 RLSDFAKSCGVPFEFHSAAMSGCE-VELENLVIRPGEALAVNFAFFLHHM-PDESVSTEN 392
RL++ A S V F F A E V+ L + P EA+AVN LH + +S +
Sbjct: 12 RLAELAXSVNVRFAFRGVAAXXXEDVKPWMLQVNPNEAVAVNSIMQLHRLLASDSDPIGS 191
Query: 393 HRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNR 452
+ +L ++SL+PK++++VEQE+N N F +RF E L YYS +++S++ DK
Sbjct: 192 GIETVLGWIRSLNPKIISVVEQEANHNQDRFLERFTEALHYYSTVFDSLEACPVEPDK-- 365
Query: 453 ISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILN 512
+ + + R+I N+++ EG RVERHE KWR R AGF P L S+ +L
Sbjct: 366 -ALAEMYLQREICNVVSSEGPARVERHEPLAKWRERLEKAGFKPLHLGSNAYKQASMLLT 542
Query: 513 EFN-ENYRLEHKDVALYLTWKNRAMCTASAWR 543
F+ E Y +E L L W +R + ASAW+
Sbjct: 543 LFSAEGYSVEENQGCLTLGWHSRPLIAASAWQ 638
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,249,304
Number of Sequences: 63676
Number of extensions: 371314
Number of successful extensions: 2162
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 2073
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2098
length of query: 545
length of database: 12,639,632
effective HSP length: 102
effective length of query: 443
effective length of database: 6,144,680
effective search space: 2722093240
effective search space used: 2722093240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0367.12


