

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.1
(481 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
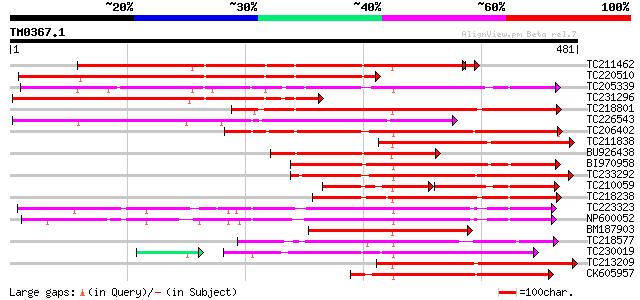
Score E
Sequences producing significant alignments: (bits) Value
TC211462 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 400 e-112
TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 362 e-100
TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucos... 331 4e-91
TC231296 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 266 2e-71
TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, ... 248 6e-66
TC226543 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransf... 238 4e-63
TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, p... 230 9e-61
TC211838 weakly similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid gluco... 212 3e-55
BU926438 similar to PIR|T03747|T03 glucosyltransferase IS5a (EC ... 202 2e-52
BI970958 197 7e-51
TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, pa... 189 2e-48
TC210059 similar to UP|Q8S9A1 (Q8S9A1) Glucosyltransferase-8 (Fr... 108 3e-45
TC218238 weakly similar to UP|Q8H0F2 (Q8H0F2) Anthocyanin 3'-glu... 173 2e-43
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 173 2e-43
NP600052 zeatin O-glucosyltransferase [Glycine max] 157 1e-38
BM187903 154 1e-37
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 151 5e-37
TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferas... 142 1e-36
TC213209 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransf... 145 3e-35
CK605957 145 4e-35
>TC211462 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (64%)
Length = 1066
Score = 400 bits (1027), Expect(2) = e-112
Identities = 197/338 (58%), Positives = 258/338 (76%), Gaps = 7/338 (2%)
Frame = +3
Query: 58 NLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVH-QGIKLLREPIQLFMEHHPADCVV 116
N +HT FPS +VGLPDG+E+ S+ DL+ +++++ LLREPI+ F+E P DC+V
Sbjct: 24 NFRVHTFDFPSEEVGLPDGVENLSAVTDLEKSYRIYIAATTLLREPIESFVERDPPDCIV 203
Query: 117 ADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKR----GSSSFLISGLPHGDIALNA 172
AD+ + W++DL +L IP L FN LF++CAM + + G F+I P + + +
Sbjct: 204 ADFLYCWVEDLAKKLRIPWLDFNGFSLFSICAMESVKKHRIGDGPFVIPDFPD-HVTIKS 380
Query: 173 SPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRG 232
+PP+ + +EPLL LKS+G +IN+F ELDG EY++HYE+TTG HKAW LGPASLVR
Sbjct: 381 TPPKDMREFLEPLLTAALKSNGFIINNFAELDGEEYLRHYEKTTG-HKAWHLGPASLVRR 557
Query: 233 TAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSV 292
T EKAERG +KSVVST E LSWL +K+ +SV+YV FGS+C F ++QL+E+ACG+EAS
Sbjct: 558 TEMEKAERG-QKSVVSTHECLSWLDSKRVNSVVYVSFGSLCYFPDKQLYEIACGMEASGY 734
Query: 293 EFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE--KGMILRGWAPQLLILGHPSVGAFLTHC 350
EF+WVVP EK+GKEEE EEEKEKWLP+GFEE KGMI++GWAPQ++IL HP+VGAFLTHC
Sbjct: 735 EFIWVVP-EKKGKEEESEEEKEKWLPKGFEERKKGMIIKGWAPQVVILEHPAVGAFLTHC 911
Query: 351 GSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVG 388
G NS +EAVSAGVP+ITWPV DQFY EKL+T+VRG+G
Sbjct: 912 GWNSTVEAVSAGVPMITWPVHSDQFYNEKLITQVRGIG 1025
Score = 25.4 bits (54), Expect(2) = e-112
Identities = 9/12 (75%), Positives = 12/12 (100%)
Frame = +1
Query: 387 VGVEVGAEEWSM 398
+GVEVGAEEW++
Sbjct: 1021 LGVEVGAEEWNL 1056
>TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (62%)
Length = 925
Score = 362 bits (928), Expect = e-100
Identities = 181/311 (58%), Positives = 236/311 (75%), Gaps = 4/311 (1%)
Frame = +3
Query: 8 RPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN---LHLHTV 64
+PLKLYFI +LA GHMIPLCD+A LF++RG HVTIITTPSN + SLP+ L LHTV
Sbjct: 3 KPLKLYFIHFLAEGHMIPLCDMATLFSTRGHHVTIITTPSNAQILRKSLPSHPLLRLHTV 182
Query: 65 PFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWI 124
FPS +VGLPDGIE+ + +DL + KV +L+ PI+ +E P DC+VADY FPW+
Sbjct: 183 QFPSHEVGLPDGIENIYADSDLDSLRKVCSATAMLQPPIEDLVEQQPPDCIVADYLFPWV 362
Query: 125 DDLTTELHIPRLSFNPSPLFALCAMRAKRGSS-SFLISGLPHGDIALNASPPEALTACVE 183
DDL +LHIPRL+FN LF +CA+ + SS S +I LPH I LNA+PP+ LT +E
Sbjct: 363 DDLAKKLHIPRLAFNGLSLFTICAIHSSSESSDSTIIQSLPH-PITLNATPPKELTKFLE 539
Query: 184 PLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHE 243
+L ELKS+G+++NSF ELDG EY ++YE+TT GHKAW LGPASL+ TA+EKAERG +
Sbjct: 540 TVLETELKSYGLIVNSFTELDGEEYTRYYEKTT-GHKAWHLGPASLIGRTAQEKAERG-Q 713
Query: 244 KSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKR 303
KSVVS E ++WL +K+ +SV+Y+CFGS+C F ++QL+E+ACGI+AS +F+WVVP EK+
Sbjct: 714 KSVVSMHECVAWLDSKRENSVVYICFGSLCYFQDKQLYEIACGIQASGHDFIWVVP-EKK 890
Query: 304 GKEEEEEEEKE 314
GKE E+EEEKE
Sbjct: 891 GKEHEKEEEKE 923
>TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose
glucosyltransferase, partial (55%)
Length = 1537
Score = 331 bits (849), Expect = 4e-91
Identities = 195/495 (39%), Positives = 286/495 (57%), Gaps = 37/495 (7%)
Frame = +2
Query: 10 LKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSL-----PNLHLHTV 64
L + P+ GH+IP+ D+A F RG TI+TTP N TI ++ ++ + TV
Sbjct: 29 LHIMLFPFPGQGHLIPMSDMARAFNGRGVRTTIVTTPLNVATIRGTIGKETETDIEILTV 208
Query: 65 PFPSRQVGLPDGIESFSS--SADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFP 122
FPS + GLP+G E+ S S DL TF + I++L P++ + H C++A FP
Sbjct: 209 KFPSAEAGLPEGCENTESIPSPDLVLTFL--KAIRMLEAPLEHLLLQHRPHCLIASAFFP 382
Query: 123 WIDDLTTELHIPRLSFNPSPLFALCAMRAKR----------GSSSFLISGLPHGDIAL-- 170
W T+L IPRL F+ + +FALCA R + F+I LP GDI +
Sbjct: 383 WASHSATKLKIPRLVFHGTGVFALCASECVRLYQPHKNVSSDTDPFIIPHLP-GDIQMTR 559
Query: 171 ---------NASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERT---TGG 218
+ LT ++ + EL S+G+++NSF EL+ Y +Y++ G
Sbjct: 560 LLLPDYAKTDGDGETGLTRVLQEIKESELASYGMIVNSFYELE-QVYADYYDKQLLQVQG 736
Query: 219 HKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNE 278
+AW +GP SL ++K +RG + SV D +L WL +KK +SV+YVCFGS+ +FS
Sbjct: 737 RRAWYIGPLSLCN---QDKGKRGKQASVDQGD-ILKWLDSKKANSVVYVCFGSIANFSET 904
Query: 279 QLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK------GMILRGWA 332
QL E+A G+E S +F+WVV +++ + WLPEGFE + G+I+ GWA
Sbjct: 905 QLREIARGLEDSGQQFIWVV--------RRSDKDDKGWLPEGFETRTTSEGRGVIIWGWA 1060
Query: 333 PQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVG 392
PQ+LIL H +VGAF+THCG NS LEAVSAGVP++TWPV +QFY EK +T++ +GV VG
Sbjct: 1061PQVLILDHQAVGAFVTHCGWNSTLEAVSAGVPMLTWPVSAEQFYNEKFVTDILQIGVPVG 1240
Query: 393 AEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGS 452
++W+ G I+ + ++KA+ R+M GEEAE +R R H+L +A A+Q GS
Sbjct: 1241VKKWNRIVGDN------ITSNALQKALHRIM-IGEEAEPMRNRAHKLAQMATTALQHNGS 1399
Query: 453 SYQNLTALIAHLKTL 467
SY + T LI HL+++
Sbjct: 1400SYCHFTHLIQHLRSI 1444
>TC231296 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (41%)
Length = 808
Score = 266 bits (680), Expect = 2e-71
Identities = 140/271 (51%), Positives = 181/271 (66%), Gaps = 7/271 (2%)
Frame = +1
Query: 3 VESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLH 62
++ +QRPLKL+FIPYL+ GH+IPLC IA LFASRGQHVT+ITTP + S P+L LH
Sbjct: 13 MDLQQRPLKLHFIPYLSPGHVIPLCGIATLFASRGQHVTVITTPYYAQILRKSSPSLQLH 192
Query: 63 TVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFP 122
V FP++ VGLPDG+E S+ DL T K +Q LLR PI FM+ HP DC+VAD +
Sbjct: 193 VVDFPAKDVGLPDGVEIKSAVTDLADTAKFYQAAMLLRRPISHFMDQHPPDCIVADTMYS 372
Query: 123 WIDDLTTELHIPRLSFNPSPLFALCAMRA-------KRGSSSFLISGLPHGDIALNASPP 175
W DD+ L IPRL+FN PLF+ AM+ + F+I PH + + + PP
Sbjct: 373 WADDVANNLRIPRLAFNGYPLFSGAAMKCVISHPELHSDTGPFVIPDFPH-RVTMPSRPP 549
Query: 176 EALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAK 235
+ TA ++ LL+ ELKSHG+++NSF ELDG E ++HYE++T GHKAW LGPA LV K
Sbjct: 550 KMATAFMDHLLKIELKSHGLIVNSFAELDGEECIQHYEKST-GHKAWHLGPACLV---GK 717
Query: 236 EKAERGHEKSVVSTDELLSWLSTKKPSSVLY 266
ERG EKSVVS +E L+WL K +SV+Y
Sbjct: 718 RDQERG-EKSVVSQNECLTWLDPKPTNSVVY 807
>TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, partial
(60%)
Length = 1281
Score = 248 bits (632), Expect = 6e-66
Identities = 134/288 (46%), Positives = 184/288 (63%), Gaps = 8/288 (2%)
Frame = +1
Query: 189 ELKSHGVLINSFVELDGH--EYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSV 246
E KS G +NSF +L+ E VK+ G KAW++GP SL TA++K ERG +
Sbjct: 73 EEKSFGTFVNSFHDLEPAYAEQVKN----KWGKKAWIIGPVSLCNRTAEDKTERG-KLPT 237
Query: 247 VSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKE 306
+ ++ L+WL++KKP+SVLYV FGS+ +EQL E+ACG+EAS F+WVV
Sbjct: 238 IDEEKCLNWLNSKKPNSVLYVSFGSLLRLPSEQLKEIACGLEASEQSFIWVVRNIHNNPS 417
Query: 307 EEEEEEKEKWLPEGFEE------KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVS 360
E +E +LPEGFE+ KG++LRGWAPQLLIL H ++ F+THCG NS LE+V
Sbjct: 418 ENKENGNGNFLPEGFEQRMKEKDKGLVLRGWAPQLLILEHVAIKGFMTHCGWNSTLESVC 597
Query: 361 AGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVR 420
AGVP+ITWP+ +QF EKL+TEV +GV+VG+ EW + ++ R+K+E AVR
Sbjct: 598 AGVPMITWPLSAEQFSNEKLITEVLKIGVQVGSREWL---SWNSEWKDLVGREKVESAVR 768
Query: 421 RLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR 468
+LM EEAE++ RV ++ A AV+ GG+SY + ALI LK R
Sbjct: 769 KLMVESEEAEEMTTRVKDIAEKAKRAVEEGGTSYADAEALIEELKARR 912
>TC226543 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransferase 73E1,
partial (21%)
Length = 1237
Score = 238 bits (607), Expect = 4e-63
Identities = 143/402 (35%), Positives = 217/402 (53%), Gaps = 24/402 (5%)
Frame = +2
Query: 3 VESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLP----- 57
+E ++ LK F+P+L+ H+IPL D+A LFA VTIITT N T S+
Sbjct: 26 MEKKKGELKSIFLPFLSTSHIIPLVDMARLFALHDVDVTIITTAHNATVFQKSIDLDASR 205
Query: 58 --NLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCV 115
+ H V FP+ QVGLP GIE+F+ + T +++ G+ LL++ + D +
Sbjct: 206 GRPIRTHVVNFPAAQVGLPVGIEAFNVDTPREMTPRIYMGLSLLQQVFEKLFHDLQPDFI 385
Query: 116 VADYCFPWIDDLTTELHIPRLSFNPSPLFALCA----------MRAKRGSSSFLISGLPH 165
V D PW D +L IPR+ F+ + A A + AK + F++ GLP
Sbjct: 386 VTDMFHPWSVDAAAKLGIPRIMFHGASYLARSAAHSVEQYAPHLEAKFDTDKFVLPGLPD 565
Query: 166 GDIALNASPPEAL------TACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGH 219
P+ L T + + + E KS+G L NSF +L+ Y +HY ++ G
Sbjct: 566 NLEMTRLQLPDWLRSPNQYTELMRTIKQSEKKSYGSLFNSFYDLES-AYYEHY-KSIMGT 739
Query: 220 KAWLLGPASL-VRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNE 278
K+W +GP SL A++KA RG+ K + L WL++K SSVLYV FGS+ F
Sbjct: 740 KSWGIGPVSLWANQDAQDKAARGYAKEEEEKEGWLKWLNSKAESSVLYVSFGSINKFPYS 919
Query: 279 QLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRGWAPQLLIL 338
QL E+A +E S +F+WVV G+ + EE EK + E KG ++ GWAPQLLIL
Sbjct: 920 QLVEIARALEDSGHDFIWVVRKNDGGEGDNFLEEFEKRMKES--NKGYLIWGWAPQLLIL 1093
Query: 339 GHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKL 380
+P++G ++HCG N+ +E+V++ +P+ TWP+ + F+ + L
Sbjct: 1094ENPAIGGLVSHCGWNTVVESVNSVLPMATWPLFAEHFFNDNL 1219
>TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (59%)
Length = 1152
Score = 230 bits (587), Expect = 9e-61
Identities = 123/294 (41%), Positives = 180/294 (60%), Gaps = 7/294 (2%)
Frame = +2
Query: 183 EPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGH 242
E + E+ S+GV++NSF EL+ Y Y++ G K W +GP SL+ +KA+RG
Sbjct: 26 EEIREAEMSSYGVVMNSFEELEP-AYATGYKKIRGD-KLWCIGPVSLINKDHLDKAQRG- 196
Query: 243 EKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEK 302
+ + + + WL +KP +V+Y C GS+C+ + QL E+ +EAS F+WV+
Sbjct: 197 -TASIDVSQHIKWLDCQKPGTVIYACLGSLCNLTTPQLKELGLALEASKRPFIWVI---- 361
Query: 303 RGKEEEEEEEKEKWLPE-GFEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALE 357
+E EE EKW+ E GFEE+ +++RGWAPQ+LIL HP++G F+THCG NS LE
Sbjct: 362 --REGGHSEELEKWIKEYGFEERTNARSLLIRGWAPQILILSHPAIGGFITHCGWNSTLE 535
Query: 358 AVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEK 417
A+ AGVP++TWP+ DQF E L+ V VGV+VG E + G+ V + + +E+
Sbjct: 536 AICAGVPMLTWPLFADQFLNESLVVHVLKVGVKVGV-EIPLTWGKEVEIGVQVKKKDVER 712
Query: 418 AVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL--KTLRD 469
A+ +LM E+E+ R+RV EL +AN AV+ GGSSY N+T LI + K RD
Sbjct: 713 AIAKLMDETSESEKRRKRVRELAEMANRAVEKGGSSYSNVTLLIQDIMQKNKRD 874
>TC211838 weakly similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid
glucosyltransferase, partial (33%)
Length = 669
Score = 212 bits (540), Expect = 3e-55
Identities = 107/171 (62%), Positives = 132/171 (76%), Gaps = 5/171 (2%)
Frame = +3
Query: 314 EKWLPEGFEE----KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWP 369
EKWLP+GFEE KGMI++GWAPQLLIL HP+VG FL+HCG NS+LEAV+AGVP+ITWP
Sbjct: 3 EKWLPKGFEERNREKGMIVKGWAPQLLILAHPAVGGFLSHCGWNSSLEAVTAGVPMITWP 182
Query: 370 VLGDQFYTEKLMTEVRGVGVEVGAEEWSM-GGGRRERERVVISRDKIEKAVRRLMGGGEE 428
V+ DQFY EKL+TEVRG+GVEVGA EW + G G RE+ +++RD IE A++RLMGGG+E
Sbjct: 183 VMADQFYNEKLITEVRGIGVEVGATEWRLVGYGEREK---LVTRDTIETAIKRLMGGGDE 353
Query: 429 AEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRDSNSVSSSMPP 479
A+ IRRR EL A ++Q GGSS+ LT LIA L LRDS S + + P
Sbjct: 354 AQNIRRRSEELAEKAKQSLQEGGSSHNRLTTLIADLMRLRDSKSAT*FLAP 506
>BU926438 similar to PIR|T03747|T03 glucosyltransferase IS5a (EC 2.4.1.-)
salicylate-induced - common tobacco, partial (21%)
Length = 446
Score = 202 bits (515), Expect = 2e-52
Identities = 100/149 (67%), Positives = 122/149 (81%), Gaps = 5/149 (3%)
Frame = +1
Query: 222 WLLGPASLVR-GTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQL 280
W LGPASL+ TA+EKAERG KS VS + +SWL +K+ +SVLY+CFGS+C F +EQL
Sbjct: 4 WHLGPASLISCRTAQEKAERGM-KSAVSMQDCVSWLDSKRVNSVLYICFGSLCHFPDEQL 180
Query: 281 HEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFE----EKGMILRGWAPQLL 336
+E+ACG+EAS EF+WVVP EK+GKE E EEEKEKWLP GFE EKGMI+RGWAPQ++
Sbjct: 181 YEIACGMEASGHEFIWVVP-EKKGKEHESEEEKEKWLPRGFEERNAEKGMIIRGWAPQVI 357
Query: 337 ILGHPSVGAFLTHCGSNSALEAVSAGVPI 365
ILGHP+VGAF+THCG NS +EAVS GVP+
Sbjct: 358 ILGHPAVGAFITHCGWNSTVEAVSEGVPM 444
>BI970958
Length = 789
Score = 197 bits (502), Expect = 7e-51
Identities = 104/235 (44%), Positives = 149/235 (63%), Gaps = 6/235 (2%)
Frame = -2
Query: 239 ERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV 298
+RG DE L WL+TK+ +SV+Y+C GS+ EQ E+A GIEAS +F+WV+
Sbjct: 782 KRGKXXEDXVDDECLKWLNTKESNSVVYICXGSLARLXKEQNXEIARGIEASGHKFLWVL 603
Query: 299 PAEKRGKEEEEEEEKEKWLPEGFEEK------GMILRGWAPQLLILGHPSVGAFLTHCGS 352
P + ++++ +E+E LP GFEE+ GM++RGW PQ LIL H ++G FLTHCG+
Sbjct: 602 P---KNTKDDDVKEEELLLPHGFEERMREKKRGMVVRGWVPQGLILKHDAIGGFLTHCGA 432
Query: 353 NSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISR 412
NS +EA+ GVP+IT P GD F EK TEV G+GVE+G EWSM +E V+
Sbjct: 431 NSVVEAICEGVPLITMPRFGDHFLCEKQATEVLGLGVELGVSEWSMSPYDARKE--VVGW 258
Query: 413 DKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTL 467
++IE AVR++M +E + +RV E+ A+ VQ GG+SY N+T L+ L+ +
Sbjct: 257 ERIENAVRKVM--KDEGGLLNKRVKEMKEKAHEVVQEGGNSYDNVTTLVQSLRRM 99
>TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, partial (46%)
Length = 993
Score = 189 bits (480), Expect = 2e-48
Identities = 104/246 (42%), Positives = 151/246 (61%), Gaps = 6/246 (2%)
Frame = +1
Query: 239 ERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV 298
+RG + S+ + L WL +K SV+YVCFGS+C+ QL E+A +E + FVWV+
Sbjct: 211 KRGDQASI-NEHHCLKWLDLQKSKSVVYVCFGSLCNLIPSQLVELALALEDTKRPFVWVI 387
Query: 299 PAEKRGKEEEEEEEKEKWLPE-GFEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSN 353
+E + +E EKW+ E GFEE+ G+I+RGWAPQ+LIL H ++G FLTHCG N
Sbjct: 388 ------REGSKYQELEKWISEEGFEERTKGRGLIIRGWAPQVLILSHHAIGGFLTHCGWN 549
Query: 354 SALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRD 413
S LE + AG+P+ITWP+ DQF EKL+T+V +GV VG E M G E+ V++ ++
Sbjct: 550 STLEGIGAGLPMITWPLFADQFLNEKLVTKVLKIGVSVGVEV-PMKFGEEEKTGVLVKKE 726
Query: 414 KIEKAVRRLM-GGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRDSNS 472
I +A+ +M GEE+++ R R +L +A AV+ GGSS+ +L+ LI + S
Sbjct: 727 DINRAICMVMDDDGEESKERRERATKLSEMAKRAVENGGSSHLDLSLLIQDIMQQSSSKE 906
Query: 473 VSSSMP 478
S P
Sbjct: 907 EIMSTP 924
>TC210059 similar to UP|Q8S9A1 (Q8S9A1) Glucosyltransferase-8 (Fragment),
partial (37%)
Length = 803
Score = 108 bits (271), Expect(2) = 3e-45
Identities = 56/100 (56%), Positives = 69/100 (69%), Gaps = 6/100 (6%)
Frame = +1
Query: 266 YVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE-- 323
Y+CFG M +FS+ QL E+A G+EAS F+WVV K+G E+ E WLPEGFEE
Sbjct: 1 YLCFGRMTAFSDAQLKEIALGLEASGQNFIWVV---KKGLNEKLE-----WLPEGFEERI 156
Query: 324 ----KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAV 359
KG+I+RGWAPQ++IL H SVG F+THCG NS LE V
Sbjct: 157 LGQGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSVLEGV 276
Score = 91.7 bits (226), Expect(2) = 3e-45
Identities = 46/106 (43%), Positives = 68/106 (63%)
Frame = +2
Query: 361 AGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVR 420
AGVP++TWP+ +QFY K +T++ +GV VG + W GR + ++ +EKAVR
Sbjct: 281 AGVPMVTWPMYSEQFYNAKFLTDIVKIGVSVGVQTWIGMMGRDP-----VKKEPVEKAVR 445
Query: 421 RLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKT 466
R+M GEEAE++R R EL +A AV+ GGSSY + +LI L++
Sbjct: 446 RIM-VGEEAEEMRNRAKELARMAKRAVEEGGSSYNDFNSLIEDLRS 580
>TC218238 weakly similar to UP|Q8H0F2 (Q8H0F2) Anthocyanin
3'-glucosyltransferase, partial (26%)
Length = 936
Score = 173 bits (438), Expect = 2e-43
Identities = 100/217 (46%), Positives = 138/217 (63%), Gaps = 6/217 (2%)
Frame = +3
Query: 258 TKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWL 317
+K +SVLYV FGSM F QL E+A +E S +F+WVV R K E E+ E +L
Sbjct: 6 SKTENSVLYVSFGSMNKFPTPQLVEIAHALEDSDHDFIWVV----RKKGESEDGEGNDFL 173
Query: 318 PEGFEE------KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVL 371
E F++ KG ++ GWAPQLLIL H ++GA +THCG N+ +E+V+AG+P+ TWP+
Sbjct: 174 QE-FDKRVKASNKGYLIWGWAPQLLILEHHAIGAVVTHCGWNTIIESVNAGLPMATWPLF 350
Query: 372 GDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQ 431
+QFY EKL+ EV +GV VGA+EW E V+ R++I A+ LM GGEE+ +
Sbjct: 351 AEQFYNEKLLAEVLRIGVPVGAKEWR---NWNEFGDEVVKREEIGNAIGVLM-GGEESIE 518
Query: 432 IRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR 468
+RRR L + A A+Q GGSS+ NL LI LK+L+
Sbjct: 519 MRRRAKALSDAAKKAIQVGGSSHNNLKELIQELKSLK 629
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 173 bits (438), Expect = 2e-43
Identities = 137/480 (28%), Positives = 222/480 (45%), Gaps = 22/480 (4%)
Frame = +1
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTID----SSLPNLHLH 62
Q + IP+ A GH+ L ++ L S V + T ++ + +S+ N+H H
Sbjct: 31 QTQVMAVLIPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFH 210
Query: 63 TVPFPSRQVGLPD-GIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADC----VVA 117
PS P+ E A L +F+ LREP++ + + V+
Sbjct: 211 AFEVPSFVSPPPNPNNEETDFPAHLLPSFEASSH---LREPVRKLLHSLSSQAKRVIVIH 381
Query: 118 DYCFPWI-DDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNASPPE 176
D + D T ++ +F+ + F + G P D L P
Sbjct: 382 DSVMASVAQDATNMPNVENYTFHSTCTFGTAVFYWDK-------MGRPLVDGMLVPEIP- 537
Query: 177 ALTACVEP------LLRKELK--SHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPAS 228
++ C + +++ + + G + N+ ++G Y++ ER TGG K W LGP +
Sbjct: 538 SMEGCFTTDFMNFMIAQRDFRKVNDGNIYNTSRAIEG-AYIEWMERFTGGKKLWALGPFN 714
Query: 229 LVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIE 288
+ K+ ER L WL + P+SVLYV FG+ +F EQ+ ++A G+E
Sbjct: 715 PLAFEKKDSKERHF---------CLEWLDKQDPNSVLYVSFGTTTTFKEEQIKKIATGLE 867
Query: 289 ASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVG 344
S +F+WV+ +G + E K FEE+ G+++R WAPQL IL H S G
Sbjct: 868 QSKQKFIWVLRDADKGDIFDGSEAKWNEFSNEFEERVEGMGLVVRDWAPQLEILSHTSTG 1047
Query: 345 AFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRE 404
F++HCG NS LE++S GVPI WP+ DQ L+TEV +G+ V + W+
Sbjct: 1048GFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGLVV--KNWA------- 1200
Query: 405 RERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
+ ++S +E AVRRLM +E + +R R L N+ + ++ GG S + + IAH+
Sbjct: 1201QRNALVSASNVENAVRRLM-ETKEGDDMRERAVRLKNVIHRSMDEGGVSRMEIDSFIAHI 1377
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 157 bits (397), Expect = 1e-38
Identities = 133/485 (27%), Positives = 223/485 (45%), Gaps = 31/485 (6%)
Frame = +1
Query: 11 KLYFIPYLAAGHMIPLCDIA---------ILFASRGQHVTIITTPSNTTTIDSSLPNLHL 61
++ IP+ A GH+ L +A + + H+ T + + I + + + H
Sbjct: 37 QVVLIPFPAQGHLNQLLHLARHIFSHNIPVHYVGTATHIRQATLRDHNSNISNIIIHFHA 216
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADC----VVA 117
VP P+ E+ S L +FK LREP++ ++ + V+
Sbjct: 217 FEVPPFVSPPPNPNNEETDFPS-HLLPSFKASSH---LREPVRNLLQSLSSQAKRVIVIH 384
Query: 118 DYCFPWI-DDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFL----ISGLPHGDIALNA 172
D + D T ++ +F+ + F ++FL + G P + A
Sbjct: 385 DSLMASVAQDATNMPNVENYTFHSTCAF-----------TTFLYYWEVMGRPPVEGFFQA 531
Query: 173 SPPEALTACVEP----LLRKELKSH----GVLINSFVELDGHEYVKHYERTTGGHKA-WL 223
+ ++ C P + +E + H G + N+ ++G Y++ ER G K W
Sbjct: 532 TEIPSMGGCFPPQFIHFITEEYEFHQFNDGNIYNTSRAIEG-PYIEFLERIGGSKKRLWA 708
Query: 224 LGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEM 283
LGP + + K+ R + WL ++ +SV+YV FG+ SF+ Q ++
Sbjct: 709 LGPFNPLTIEKKDPKTR---------HICIEWLDKQEANSVMYVSFGTTTSFTVAQFEQI 861
Query: 284 ACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILG 339
A G+E S +F+WV+ +G + E + LP GFEE+ G+++R WAPQL IL
Sbjct: 862 AIGLEQSKQKFIWVLRDADKGNIFDGSEAERYELPNGFEERVEGIGLLIRDWAPQLEILS 1041
Query: 340 HPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMG 399
H S G F++HCG NS LE+++ GVPI WP+ DQ L+TEV VG V ++W+
Sbjct: 1042HTSTGGFMSHCGWNSCLESITMGVPIAAWPMHSDQPRNSVLITEVLKVGFVV--KDWA-- 1209
Query: 400 GGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTA 459
+ ++S +E AVRRLM +E +++R R L N + + GG S + +
Sbjct: 1210-----QRNALVSASVVENAVRRLM-ETKEGDEMRDRAVRLKNCIHRSKYGGGVSRMEMGS 1371
Query: 460 LIAHL 464
IAH+
Sbjct: 1372FIAHI 1386
>BM187903
Length = 438
Score = 154 bits (388), Expect = 1e-37
Identities = 76/145 (52%), Positives = 100/145 (68%), Gaps = 6/145 (4%)
Frame = +3
Query: 254 SWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEK 313
+WL++KKP+SVLYV FGS+ QL E+A G+EAS F+WVV + E +E
Sbjct: 3 NWLNSKKPNSVLYVSFGSVARLPPGQLKEIAFGLEASDQTFIWVVGCIRNNPSENKENGS 182
Query: 314 EKWLPEGFEE------KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIIT 367
+L EGFE+ KG++LRGWAPQLLIL H ++ F+THCG NS LE+V AGVP+IT
Sbjct: 183 GNFLFEGFEQRMKEKNKGLVLRGWAPQLLILEHAAIKGFMTHCGWNSTLESVCAGVPMIT 362
Query: 368 WPVLGDQFYTEKLMTEVRGVGVEVG 392
WP+ +QF EKL+TEV +GV+VG
Sbjct: 363 WPLSAEQFSNEKLITEVLKIGVQVG 437
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 151 bits (382), Expect = 5e-37
Identities = 98/280 (35%), Positives = 149/280 (53%), Gaps = 8/280 (2%)
Frame = +1
Query: 194 GVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELL 253
GV +N+F+EL+ E G K + +GP + GHE V E L
Sbjct: 607 GVFMNTFLELESGAIRALEEHVKGKPKLYPVGPIIQMESI-------GHENGV----ECL 753
Query: 254 SWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEK----RGKEEEE 309
+WL ++P+SVLYV FGS + S EQ +E+A G+E S +F+WVV A G E
Sbjct: 754 TWLDKQEPNSVLYVSFGSGGTLSQEQFNELAFGLELSGKKFLWVVRAPSGVVSAGYLCAE 933
Query: 310 EEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPI 365
++ ++LP GF E+ G+++ WAPQ+ +LGH + G FL+HCG NS LE+V GVP+
Sbjct: 934 TKDPLEFLPHGFLERTKKQGLVVPSWAPQIQVLGHSATGGFLSHCGWNSVLESVVQGVPV 1113
Query: 366 ITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGG 425
ITWP+ +Q ++ A++ + + E ++ R++I K VR LMG
Sbjct: 1114ITWPLFAEQSLNAAMI-----------ADDLKVALRPKVNESGLVEREEIAKVVRGLMGD 1260
Query: 426 GEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLK 465
E E IR+R+ L A A++ GSS + L+ + L+
Sbjct: 1261KESLE-IRKRMGLLKIAAANAIKEDGSSTKTLSEMATSLR 1377
>TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferase-like
protein , partial (36%)
Length = 1202
Score = 142 bits (359), Expect(2) = 1e-36
Identities = 88/275 (32%), Positives = 148/275 (53%), Gaps = 8/275 (2%)
Frame = +2
Query: 182 VEPLLRKELKSHGVLINSFVELD--GHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAE 239
+ P + +KS G + N+ E++ G + +++Y + LL PASL+ +
Sbjct: 284 IVPQIALSMKSDGWICNTVQEIEPLGLQLLRNYLQLPVWPVGPLLPPASLM-----DSKH 448
Query: 240 RGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVP 299
R ++S ++ D + WL +K SSVLY+ FGS + + Q+ +A G+E S F+W++
Sbjct: 449 RAGKESGIALDACMQWLDSKDESSVLYISFGSQNTITASQMMALAEGLEESGRSFIWII- 625
Query: 300 AEKRGKEEEEEEEKEKWLPEGFEEK------GMILRGWAPQLLILGHPSVGAFLTHCGSN 353
G + E E WLP+GFEE+ G+++ W PQL IL H S GAFL+HCG N
Sbjct: 626 RPPFGFDINGEFIAE-WLPKGFEERMRDTKRGLLVNKWGPQLEILSHSSTGAFLSHCGWN 802
Query: 354 SALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRD 413
S LE++S GVP+I WP+ +Q Y K++ E GV +E+ ++ G++ ++ + I+ +
Sbjct: 803 SVLESLSYGVPMIGWPLAAEQTYNLKMLVEEMGVAIELTQTVETVISGKQVKKVIEIAME 982
Query: 414 KIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQ 448
+ K E A +R + E G ++V+
Sbjct: 983 QEGKGKEMKEKANEIAAHMREAITEKGKEKGSSVR 1087
Score = 28.9 bits (63), Expect(2) = 1e-36
Identities = 17/65 (26%), Positives = 25/65 (38%), Gaps = 8/65 (12%)
Frame = +3
Query: 108 EHHPADCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAM--------RAKRGSSSFL 159
E HP C ++D W++++ L I LSF + A K S F
Sbjct: 9 EGHPPLCTISDVFLGWVNNVAKSLCIRNLSFTTCGAYGTLAYVSIWFNLPHRKTDSDEFC 188
Query: 160 ISGLP 164
G+P
Sbjct: 189 APGVP 203
>TC213209 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransferase 73E1,
partial (25%)
Length = 624
Score = 145 bits (367), Expect = 3e-35
Identities = 78/175 (44%), Positives = 108/175 (61%), Gaps = 5/175 (2%)
Frame = +3
Query: 312 EKEKWLPE-GFEE----KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPII 366
E EKWL E F+E KG+I+RGWAPQ+ IL HPS G FL+HCG NS LEAVSAG+P+I
Sbjct: 3 EIEKWLEEENFQERNRRKGVIIRGWAPQVEILSHPSTGGFLSHCGWNSTLEAVSAGIPMI 182
Query: 367 TWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGG 426
TWP+ +QF EKL+ +V +GV +G E E ++ ++ ++ ++KAV +LM G
Sbjct: 183 TWPMSAEQFINEKLIVQVLKIGVRIGVE---APVDPMETQKALVKKECVKKAVDQLMEQG 353
Query: 427 EEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLRDSNSVSSSMPPLH 481
+ EQ R R E+ +A AV+ GGSS N I + + D S +S P +H
Sbjct: 354 GDGEQRRNRAREIKEMAQKAVEDGGSSASNCELFIQEIGAVEDGGSSASK*P*VH 518
>CK605957
Length = 648
Score = 145 bits (366), Expect = 4e-35
Identities = 78/177 (44%), Positives = 113/177 (63%), Gaps = 5/177 (2%)
Frame = -2
Query: 290 SSVEFVWVVPAEKRGKEEEEEEEKEKWLP-EGFEEK----GMILRGWAPQLLILGHPSVG 344
S+ F+WVV E E EKWL E FEE+ G++++GWAPQ+LIL HPS+G
Sbjct: 647 SNRPFIWVVKTIG-----ENFSEVEKWLEDENFEERVKGRGLLIKGWAPQILILSHPSIG 483
Query: 345 AFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRE 404
FLTHCG NS +E+V +GVP+ITWP+ +QF EK + EV +GV +G E + G +
Sbjct: 482 GFLTHCGWNSTIESVCSGVPMITWPLFAEQFLNEKCIVEVLKIGVRIGVEV-PVRFGDEK 306
Query: 405 RERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
+ V++ + +I +A+ +M GGEE ++ R V ELGN+A A++ GSS N++ LI
Sbjct: 305 KGGVLVKKIQIMEAIEMIMEGGEEGDKRRSGVTELGNIARRALEEEGSSRFNISCLI 135
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,509,049
Number of Sequences: 63676
Number of extensions: 367512
Number of successful extensions: 3220
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 2619
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2833
length of query: 481
length of database: 12,639,632
effective HSP length: 101
effective length of query: 380
effective length of database: 6,208,356
effective search space: 2359175280
effective search space used: 2359175280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0367.1


