

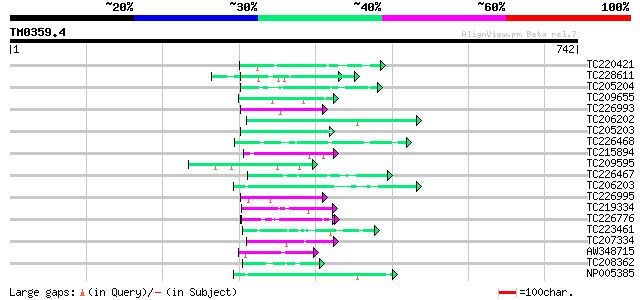
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0359.4
(742 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 57 3e-08
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 49 8e-06
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 48 2e-05
TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partia... 48 2e-05
TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 deh... 48 2e-05
TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragme... 47 4e-05
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 47 4e-05
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 46 5e-05
TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase ... 45 2e-04
TC209595 homologue to GB|AAL62011.1|18252261|AY072620 AT5g08330/... 45 2e-04
TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 44 3e-04
TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragme... 44 3e-04
TC226995 similar to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehyd... 43 4e-04
TC219334 weakly similar to UP|Q9FUZ9 (Q9FUZ9) SKP1 interacting p... 43 4e-04
TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {A... 43 4e-04
TC223461 similar to UP|C941_VICSA (O81117) Cytochrome P450 94A1 ... 43 4e-04
TC207334 similar to UP|Q949K3 (Q949K3) Suppressor-like protein, ... 43 6e-04
AW348715 43 6e-04
TC208362 weakly similar to UP|O64835 (O64835) Expressed protein ... 43 6e-04
NP005385 DNA-directed RNA polymerase 43 6e-04
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 57.0 bits (136), Expect = 3e-08
Identities = 56/196 (28%), Positives = 72/196 (36%), Gaps = 4/196 (2%)
Frame = +3
Query: 301 HSSNPLTPIPESQPAAQTTSPPH---SPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLL 357
H +PL + QP +TSPPH +P P P + +P S PT+
Sbjct: 156 HHPHPLWTQNKQQPLNPSTSPPHRTPAPSPPSTMPPSATPPSPSATSSP---SASPTAPP 326
Query: 358 TIPYDPLSSEPIVQDQPEPNQTEPQ-PRTSDPSAPRASERPAARTTDTDSSTAFTPVSFP 416
T P S P P P+ T P P S P++P S A T +S + +P S
Sbjct: 327 TSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTS-- 500
Query: 417 INVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPD 476
+ PS TS S K A PS + P P PI PD
Sbjct: 501 -RTSPTSPSPTSTS----------KPPAPSSSSPA*PSSKPSPSP---------TPISPD 620
Query: 477 PEPAQSVSNQSSVRSP 492
P PA S + SP
Sbjct: 621 PSPATSTPTSHTSISP 668
Score = 41.2 bits (95), Expect = 0.002
Identities = 44/160 (27%), Positives = 65/160 (40%), Gaps = 8/160 (5%)
Frame = +3
Query: 263 TEPPARVTRSSAHSSKPALA---SDDDLNLFDALPISALLQHS-SNPLTPIPESQPAAQT 318
T PP R S S+ P A S + A P + S S P +P P S P+ +
Sbjct: 210 TSPPHRTPAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSS 389
Query: 319 T--SPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEP 376
T SPP + + PSP +P S S + T P +S+P P P
Sbjct: 390 TAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKP-----PAP 554
Query: 377 NQTEPQPRTS--DPSAPRASERPAARTTDTDSSTAFTPVS 414
+ + P +S PS S P+ T+ S T+ +P++
Sbjct: 555 SSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSISPIT 674
Score = 39.3 bits (90), Expect = 0.006
Identities = 42/149 (28%), Positives = 58/149 (38%), Gaps = 8/149 (5%)
Frame = +3
Query: 250 PTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPI 309
P PA + P A SA SS A + + + P S + +P +
Sbjct: 216 PPHRTPAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTA 395
Query: 310 PESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSR----SEDPTSLLTIPYDPLS 365
P A+ TSP SP S P+ T + +PTSR S PTS P S
Sbjct: 396 PSPPSASPPTSPSPSPPS----PASTASANSPASGSPTSRTSPTSPSPTSTSKPPAPSSS 563
Query: 366 SEPIVQDQPEPNQT----EPQPRTSDPSA 390
S +P P+ T +P P TS P++
Sbjct: 564 SPA*PSSKPSPSPTPISPDPSPATSTPTS 650
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 48.9 bits (115), Expect = 8e-06
Identities = 50/164 (30%), Positives = 64/164 (38%), Gaps = 8/164 (4%)
Frame = +3
Query: 302 SSNPLTPIPESQPAAQTTSPPHS---PRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLT 358
S P P P S ++ T S P + P SS +PSP + PTS S P + T
Sbjct: 165 SPAPPPPPPRSASSSTTASSPTAVTPPSSSPPRPSPGPSA------TPTSTSPSPATPST 326
Query: 359 ---IPYDPLSSEPIVQDQPEPNQTEPQPRTSDP--SAPRASERPAARTTDTDSSTAFTPV 413
P PLSS P P P + P P TS P +AP + AA T + S + P
Sbjct: 327 SSSAPRPPLSS-PAPTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEPPSPPPYPPT 503
Query: 414 SFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRR 457
S P N+S + + V LT PS R
Sbjct: 504 SPPPPCPSLKSPNSSPNAASPSKNSSPSVGPTPSDSLTAPSSSR 635
Score = 45.8 bits (107), Expect = 7e-05
Identities = 47/183 (25%), Positives = 68/183 (36%), Gaps = 10/183 (5%)
Frame = +3
Query: 265 PPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNP-LTPIPESQPAAQTTSPPH 323
PP R SS +S P + P S+ + S P TP S A ++
Sbjct: 183 PPPRSASSSTTASSPTAVTP---------PSSSPPRPSPGPSATPTSTSPSPATPSTSSS 335
Query: 324 SPRSSFFQPSPTEAPLWNLLQNPTSRS-------EDPTSLLTIPYDPLSSEPIVQDQPEP 376
+PR P+PT +P P + S P+S +P S P P P
Sbjct: 336 APRPPLSSPAPTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEPPSPPPYPPTSPPP 515
Query: 377 NQTEPQPRTSDPSA--PRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKF 434
+ S P+A P + P+ T +DS TA P S + PP T ++R F
Sbjct: 516 PCPSLKSPNSSPNAASPSKNSSPSVGPTPSDSLTA--PSSSRTSPTTLPPPTTLATLRGF 689
Query: 435 MEI 437
+
Sbjct: 690 KRL 698
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 47.8 bits (112), Expect = 2e-05
Identities = 53/187 (28%), Positives = 69/187 (36%), Gaps = 1/187 (0%)
Frame = +1
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPY 361
S++P P P+S P +SPP P S P P ++P P S T P
Sbjct: 187 STSPAPPTPQSSPPPIQSSPPPVPSS----PPPAQSP-------------PPAS--TPPP 309
Query: 362 DPLSSEPIVQDQP-EPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVI 420
PLSS P P P P P + P++P + P A A P + P
Sbjct: 310 APLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATPPPASPPPF- 486
Query: 421 DSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPA 480
SPP T AL L+ P P P P P ++V P + P P
Sbjct: 487 -SPPPATPPP--------ATPPPALTPTPLSSP-PATSPAPAPAKVVAP--ALSPSLAPG 630
Query: 481 QSVSNQS 487
S+S S
Sbjct: 631 PSLSTIS 651
Score = 39.3 bits (90), Expect = 0.006
Identities = 41/166 (24%), Positives = 54/166 (31%)
Frame = +1
Query: 255 PAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQP 314
PA + + PP + + SS P S + P+S+ S P +P P S P
Sbjct: 196 PAPPTPQSSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPASPPPSSPPPASPP 375
Query: 315 AAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQP 374
A + PP SP + P P P P S P
Sbjct: 376 PA--SPPPASPPPA--SPPPASPP------------------------PASPPPATPPPA 471
Query: 375 EPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVI 420
P P P T P+ P PA T S A +P P V+
Sbjct: 472 SPPPFSPPPATPPPATP----PPALTPTPLSSPPATSPAPAPAKVV 597
Score = 31.2 bits (69), Expect = 1.7
Identities = 27/103 (26%), Positives = 40/103 (38%), Gaps = 1/103 (0%)
Frame = +1
Query: 235 PPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALP 294
PP + P P ++ + PPA +S + P AS + A P
Sbjct: 283 PPPASTPPPAPLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATP 462
Query: 295 ISAL-LQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTE 336
A S P TP P + P A T +P SP ++ P+P +
Sbjct: 463 PPASPPPFSPPPATPPPATPPPALTPTPLSSPPATSPAPAPAK 591
>TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partial (32%)
Length = 832
Score = 47.8 bits (112), Expect = 2e-05
Identities = 40/137 (29%), Positives = 55/137 (39%), Gaps = 6/137 (4%)
Frame = +1
Query: 300 QHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNL---LQNPTSRSEDPTSL 356
Q SS A TTSPP SP S +P PT P L P+S S PT+
Sbjct: 13 QSSSKKAPTTSSPSTTAATTSPPSSPSSPTHRPLPTRTPTQPLPPHSAQPSSNSPSPTAP 192
Query: 357 LTIPYDPLSSEPIVQDQPEPNQTEPQ---PRTSDPSAPRASERPAARTTDTDSSTAFTPV 413
+ SS P P P R + ++P +S P++ T ++ P
Sbjct: 193 SRLHSGTRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPSAAPKSSTKP- 369
Query: 414 SFPINVIDSPPSNTSES 430
S+ +V S PS T+ S
Sbjct: 370 SYSPSVRSSLPSPTTRS 420
Score = 34.7 bits (78), Expect = 0.16
Identities = 46/175 (26%), Positives = 67/175 (38%), Gaps = 2/175 (1%)
Frame = +1
Query: 235 PPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALP 294
PP + KK PT P+ T PP+ + S H P P
Sbjct: 4 PPNQSSSKKA-----PTTSSPSTTAAT-TSPPSSPS-SPTHRPLPTRT-----------P 129
Query: 295 ISALLQHSSNPL--TPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSED 352
L HS+ P +P P + + + SP + PSP+ P ++ +N S S
Sbjct: 130 TQPLPPHSAQPSSNSPSPTAPSRLHSGTRSSSPSAPPPPPSPSRTPSFHGSRNTISAS-- 303
Query: 353 PTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSS 407
PTS T S+ P +P P R+S PS + R +AR +T +S
Sbjct: 304 PTSSSTPSSSTPSAAPKSSTKP---SYSPSVRSSLPS---PTTRSSARAPETATS 450
Score = 30.8 bits (68), Expect = 2.3
Identities = 27/82 (32%), Positives = 35/82 (41%), Gaps = 2/82 (2%)
Frame = +1
Query: 270 TRSSAHSSKPALASDDDLNLFDAL--PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRS 327
TRSS+ S+ P S F ISA SS P + P + P + +T P +SP
Sbjct: 211 TRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPSAAPKS-STKPSYSPSV 387
Query: 328 SFFQPSPTEAPLWNLLQNPTSR 349
PSPT + TSR
Sbjct: 388 RSSLPSPTTRSSARAPETATSR 453
>TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (94%)
Length = 1388
Score = 47.8 bits (112), Expect = 2e-05
Identities = 39/132 (29%), Positives = 56/132 (41%), Gaps = 17/132 (12%)
Frame = +2
Query: 302 SSNPLTPIPESQPAAQT-TSPPHSPRS--SFFQPSPTEAPLWNLLQNPTSRSED------ 352
S+ P++P P A T +SP S S S PSP+ P +P + S
Sbjct: 314 STTPISPTPPPSAAGSTPSSPTRSTTSPLSLTSPSPSRYPTTPPTSSPPAPSASSRPSVP 493
Query: 353 -------PTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSA-PRASERPAARTTDT 404
PTS T P P S P ++ P+P + P P T P+A P + AR T +
Sbjct: 494 TSPPPAAPTSATTKPAPPRCSAPPLRRSPKPPPSTPAPPTPPPNAPPTGTP*TTARLTPS 673
Query: 405 DSSTAFTPVSFP 416
+TA + + P
Sbjct: 674 SPATASSSTTSP 709
Score = 37.0 bits (84), Expect = 0.032
Identities = 40/139 (28%), Positives = 50/139 (35%), Gaps = 17/139 (12%)
Frame = +2
Query: 390 APRASERPAARTTDTDSSTAFTPV-SFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEY 448
APR S R A+ T+ +T P S + +PP + + S S
Sbjct: 236 APRTSTRSASTTSMWIPTTPTRPA*SSTTPISPTPPPSAAGSTPS-----SPTRSTTSPL 400
Query: 449 YLTCPSPRRYP-------GPRPERLVDPDEPIQPDPEPAQSVSNQSSV---------RSP 492
LT PSP RYP P P P P P P S + + + RSP
Sbjct: 401 SLTSPSPSRYPTTPPTSSPPAPSASSRPSVPTSPPPAAPTSATTKPAPPRCSAPPLRRSP 580
Query: 493 HPLVETSDPHLGTSDPNAP 511
P T P T PNAP
Sbjct: 581 KPPPSTPAP--PTPPPNAP 631
>TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragment) ,
complete
Length = 1826
Score = 46.6 bits (109), Expect = 4e-05
Identities = 60/238 (25%), Positives = 85/238 (35%), Gaps = 9/238 (3%)
Frame = +3
Query: 310 PESQPAAQTTSPPHSPRSSFFQP-SPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEP 368
P S P +SP +SP S + P SP +P + +PTS PTS P P S
Sbjct: 612 PTSSPGYSPSSPGYSPSSPGYSPTSPGYSPT-SPGYSPTSPGYSPTSPTYSPSSPGYSPT 788
Query: 369 IVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPS-NT 427
P P + P++P S + + + S + +P P + SP S +
Sbjct: 789 SPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPAYSPTSPSY 968
Query: 428 SESIRKFMEIRKEKVSALEEYYLTCP-----SPRRYPGPRPERLVDPD-EPIQPDPEP-A 480
S + + Y T P SP P P P P P +
Sbjct: 969 SPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPGYSPTSPSYSPTSPSYSPTS 1148
Query: 481 QSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHLASPEPNLS 538
S + QS+ SP S P L S P +P SP S + +S + P S
Sbjct: 1149PSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYS 1322
Score = 32.7 bits (73), Expect = 0.60
Identities = 31/112 (27%), Positives = 44/112 (38%), Gaps = 1/112 (0%)
Frame = +3
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPY 361
S +P +P Q A + S +SP S PS +P + +PTS S PTS P
Sbjct: 1131 SYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPT-SPNYSPTSPSYSPTSPSYSPS 1307
Query: 362 DPLSSEPIVQDQPEPNQTEPQPRTSDPS-APRASERPAARTTDTDSSTAFTP 412
P S P + P S P +P P+ S++ +TP
Sbjct: 1308 SPTYS----PSSPYNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYTP 1451
Score = 31.6 bits (70), Expect = 1.3
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 1/94 (1%)
Frame = +3
Query: 304 NPLTPIPESQPAAQTTSPPHSPRSSFFQP-SPTEAPLWNLLQNPTSRSEDPTSLLTIPYD 362
+P +P + P TSP +SP S + P SPT +P +P + P Y
Sbjct: 1218 SPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSP-----SSPYNSGVSP------DYS 1364
Query: 363 PLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASER 396
P S + P+Q P ++ P+ S++
Sbjct: 1365 PSSPQFSPSTGYSPSQPGYSPSSTSQYTPQTSDK 1466
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 46.6 bits (109), Expect = 4e-05
Identities = 34/125 (27%), Positives = 49/125 (39%), Gaps = 2/125 (1%)
Frame = +3
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPRS--SFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTI 359
S++P P P++ P +SPP P S P P P + L +P S P+S
Sbjct: 279 STSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPS 458
Query: 360 PYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINV 419
P S P P + P P + P +P + P A + TP P++
Sbjct: 459 SPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLS- 635
Query: 420 IDSPP 424
SPP
Sbjct: 636 --SPP 644
Score = 40.8 bits (94), Expect = 0.002
Identities = 42/145 (28%), Positives = 56/145 (37%), Gaps = 5/145 (3%)
Frame = +3
Query: 255 PAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQP 314
PA + + PP + + SS P S + P+S+ S P +P P S P
Sbjct: 288 PAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPP 467
Query: 315 AAQ---TTSPPHSPRSSFFQPS--PTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPI 369
A ++ PP SP S PS P +P T P LT PLSS P+
Sbjct: 468 PASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPV 647
Query: 370 VQDQPEPNQTEPQPRTSDPSAPRAS 394
P P++ P S AP S
Sbjct: 648 SSPAPSPSKVF-APALSPSLAPGPS 719
Score = 33.5 bits (75), Expect = 0.35
Identities = 46/192 (23%), Positives = 62/192 (31%), Gaps = 10/192 (5%)
Frame = +3
Query: 339 LWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDP---------- 388
++ L+ ++++ P S P P +S P VQ P P + P P S P
Sbjct: 234 IFTLVAGVGAQAQGP-STSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPL 410
Query: 389 SAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEY 448
S+P S P + + A P S P P S S F
Sbjct: 411 SSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPPF-------------- 548
Query: 449 YLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDP 508
P P P P V P + P P S S SP + P L S
Sbjct: 549 ---SPPPATPPPATPPPAVPPPS-LTPTVTPLSSPPVSSPAPSPSKVFA---PALSPSLA 707
Query: 509 NAPMINIGSPQG 520
P ++ SP G
Sbjct: 708 PGPSLSTISPSG 743
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 46.2 bits (108), Expect = 5e-05
Identities = 52/232 (22%), Positives = 83/232 (35%), Gaps = 1/232 (0%)
Frame = -2
Query: 295 ISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPT 354
IS + N P TT PP +P ++ SP+ + + Q PT + PT
Sbjct: 1019 ISLCCKLDCNAQAPATVPVKLPPTTLPPTTPTAT----SPSSSTPLAVTQPPTVVASPPT 852
Query: 355 SLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVS 414
+ S P QP P P+P PSAP+ + + + P
Sbjct: 851 T----------STPPTTSQP-PANVAPKPAPVKPSAPKVAPASSPKAPPPQIPIPQPPKP 705
Query: 415 FPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRY-PGPRPERLVDPDEPI 473
P+ SPP + + K++ P+P + P P P P
Sbjct: 704 SPV----SPPPLPPPPVASPPPLPPPKIAPTPAKTPPSPAPAKATPAPAPA-------PT 558
Query: 474 QPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAH 525
+P P P+ ++ +P P++E P AP+I + +P A H
Sbjct: 557 KPAPTPSPVPPPPAATPAPAPVIEVPAP------APAPVIEVPAPAPAHHRH 420
Score = 41.6 bits (96), Expect = 0.001
Identities = 58/249 (23%), Positives = 92/249 (36%), Gaps = 18/249 (7%)
Frame = -2
Query: 263 TEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPP 322
T PP T +S SS P LA + + P ++ +S P + + ++P
Sbjct: 947 TLPPTTPTATSPSSSTP-LAVTQPPTVVASPPTTSTPPTTSQPPANVAPKPAPVKPSAPK 771
Query: 323 HSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQ---PEPNQT 379
+P SS P P + P+ P P S +P P++S P + P P +T
Sbjct: 770 VAPASSPKAPPP-QIPI------PQPPKPSPVSPPPLPPPPVASPPPLPPPKIAPTPAKT 612
Query: 380 EPQPRTSD--PSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSP----------PSNT 427
P P + P+ A +PA + A TP P+ + +P P+
Sbjct: 611 PPSPAPAKATPAPAPAPTKPAPTPSPVPPPPAATPAPAPVIEVPAPAPAPVIEVPAPAPA 432
Query: 428 SESIRKFMEIRKEKVSALEEYYLTCPSPR--RYPGPRPERLVDPDEPIQPDPEPAQSVS- 484
R+ + + + P+P R P P PD + D PA S S
Sbjct: 431 HHRHRRHRHRHRHR-----RHQAPAPAPTIIRKSPPAP-----PDSTAESDTAPAPSPSL 282
Query: 485 NQSSVRSPH 493
N ++ S H
Sbjct: 281 NLNAAPSNH 255
>TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase , complete
Length = 1675
Score = 44.7 bits (104), Expect = 2e-04
Identities = 45/139 (32%), Positives = 62/139 (44%), Gaps = 15/139 (10%)
Frame = +1
Query: 307 TPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPL-WNL-LQNPTSRSEDPTSLLTIPYDPL 364
+P P S P T+PP PR S SP+ AP W + P+S PTS + P
Sbjct: 208 SPRPSSGP*---TTPPLPPRPSATTRSPSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPT 378
Query: 365 SSEPIVQDQPEPNQTEPQPRTSDPSAP-------RASERPA-ARTTDTDSSTA---FTPV 413
SS P + P PR++ P P AS PA AR+ + +ST+ +P
Sbjct: 379 SSAARCSTSSTPPPSSPAPRSTPPPTPPSPPAPTSASSPPAPARSPASHASTSSRGTSPS 558
Query: 414 SFPINVIDS--PPSNTSES 430
S P+ + S PP+ S S
Sbjct: 559 SAPLFPLSSVTPPTQLSSS 615
Score = 39.7 bits (91), Expect = 0.005
Identities = 39/151 (25%), Positives = 62/151 (40%), Gaps = 2/151 (1%)
Frame = +1
Query: 379 TEPQPRTSDPSAPRASERPAARTTDTDSSTAFT--PVSFPINVIDSPPSNTSESIRKFME 436
T P+P + + P RP+A T S+ A P P ++ SP S++S +
Sbjct: 205 TSPRPSSGP*TTPPLPPRPSATTRSPSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPTSS 384
Query: 437 IRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLV 496
+ S+ + P+PR P P P P P P PA+S ++ +S S
Sbjct: 385 AARCSTSSTPPP--SSPAPRSTPPPTPPSPPAPTSASSP-PAPARSPASHASTSSR---- 543
Query: 497 ETSDPHLGTSDPNAPMINIGSPQGASEAHSS 527
GTS +AP+ + S ++ SS
Sbjct: 544 -------GTSPSSAPLFPLSSVTPPTQLSSS 615
Score = 35.8 bits (81), Expect = 0.071
Identities = 35/133 (26%), Positives = 52/133 (38%), Gaps = 1/133 (0%)
Frame = +1
Query: 229 SESEDGPPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALA-SDDDL 287
S S PP + P+ P +A + PPA ++H+S + S
Sbjct: 397 STSSTPPPSSPAPRSTPPPTPPSPPAPTSA----SSPPAPARSPASHASTSSRGTSPSSA 564
Query: 288 NLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPT 347
LF ++ Q SS+ P P + S P SP ++ P+PT W L P
Sbjct: 565 PLFPLSSVTPPTQLSSS--FPTPSTSSPTSRGSSPASPPTASSAPAPT----WTL---PA 717
Query: 348 SRSEDPTSLLTIP 360
S S P +L + P
Sbjct: 718 SVSSSPITLTSTP 756
>TC209595 homologue to GB|AAL62011.1|18252261|AY072620 AT5g08330/F8L15_60
{Arabidopsis thaliana;} , partial (56%)
Length = 869
Score = 44.7 bits (104), Expect = 2e-04
Identities = 51/201 (25%), Positives = 71/201 (34%), Gaps = 33/201 (16%)
Frame = +2
Query: 235 PPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPA----RVTRSSAHSSKPALASDDDLN-- 288
PP + + R + V AA R + PPA R + +++ +++P+ S N
Sbjct: 71 PPSPSRNRLPRTATARSTVAGAAFACRSSAPPASSSSRASSATSPTARPSSGSSARRNPP 250
Query: 289 ---------LFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPL 339
L A P+S SS PL P + A TT+PP S P P P
Sbjct: 251 SSPLPAPAPLRRASPLSPTTSPSSLPLPPSSSANAFALTTTPPPKTTPSRSSPPPRRPPS 430
Query: 340 WNLLQNPTS---------------RSEDPTSLLTIPYDPLSSEPIVQDQPEPNQ---TEP 381
L PTS R +TI +S ++P P T P
Sbjct: 431 GPYLPAPTSARSGASPPRPNSCRCRRRIQCFTITINTTSSNSNRPPWERPLPQGSGITSP 610
Query: 382 QPRTSDPSAPRASERPAARTT 402
P P A+ PA TT
Sbjct: 611 ATSICSPRCPAATGIPAGGTT 673
>TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 1017
Score = 43.9 bits (102), Expect = 3e-04
Identities = 48/193 (24%), Positives = 74/193 (37%), Gaps = 3/193 (1%)
Frame = +3
Query: 312 SQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEP--I 369
S A T+P +SP ++ SP+ + PT + PT+ P +S+P I
Sbjct: 39 SNAQAPATAPANSPPTTPAATSPSSGTPLAVTLPPTVVASPPTTSTP----PTTSQPPAI 206
Query: 370 VQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSE 429
V +P P T P P+ + S+P+A + S TP S P I PP +T
Sbjct: 207 VAPKPAP-VTPPAPKVAPASSPKAPPPQTPQPQPPKVSPVVTPTSPP--PIPPPPVSTPP 377
Query: 430 SIRKFMEIRKEKVSALEEYYLTCPSPRRYP-GPRPERLVDPDEPIQPDPEPAQSVSNQSS 488
+ K++ P+P + P P P + P P P +
Sbjct: 378 PLPP------PKIA---------PTPAKTPPAPAPAKATPAPAPAPTKPAPTPAPVPPPP 512
Query: 489 VRSPHPLVETSDP 501
+P P+VE P
Sbjct: 513 TLAPTPVVEVPAP 551
Score = 29.3 bits (64), Expect = 6.6
Identities = 51/257 (19%), Positives = 77/257 (29%), Gaps = 13/257 (5%)
Frame = +3
Query: 250 PTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFD----ALPISALLQHSSNP 305
P P++ P V S +S P S + P + + +S+P
Sbjct: 90 PAATSPSSGTPLAVTLPPTVVASPPTTSTPPTTSQPPAIVAPKPAPVTPPAPKVAPASSP 269
Query: 306 LTPIPES--------QPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLL 357
P P++ P TSPP P P P P
Sbjct: 270 KAPPPQTPQPQPPKVSPVVTPTSPPPIP------PPPVSTP------------------- 374
Query: 358 TIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPI 417
P P + P P P + P+ A +PA P P+
Sbjct: 375 -----PPLPPPKIAPTPAKTPPAPAPAKATPAPAPAPTKPAPTPAPVPPPPTLAPT--PV 533
Query: 418 NVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDP 477
+ +P ++ + R R+ + A P+ R P P PD + D
Sbjct: 534 VEVPAPAPSSHKHRRHRHRHRRHQAPA------PAPTIVRKSPPAP-----PDSTAESDT 680
Query: 478 EPAQSVS-NQSSVRSPH 493
PA S S N ++ S H
Sbjct: 681 TPAPSPSLNLNAASSNH 731
>TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragment) , partial
(96%)
Length = 2604
Score = 43.5 bits (101), Expect = 3e-04
Identities = 61/246 (24%), Positives = 84/246 (33%), Gaps = 1/246 (0%)
Frame = +1
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQP-SPTEAPLWNLLQNPTSRSED 352
P S SS +P + PA TSP +SP S + P SP+ +P + +PTS S
Sbjct: 1198 PTSPTYSPSSPGYSP---TSPAYSPTSPSYSPTSPSYSPTSPSYSPT-SPSYSPTSPSYS 1365
Query: 353 PTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTP 412
PTS P P S P P + P++P S + + + S + +P
Sbjct: 1366 PTSPAYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSP 1545
Query: 413 VSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEP 472
P + SP S Y T PS P P
Sbjct: 1546 AYSPTSPGYSPTS--------------------PSYSPTSPS------------YSPTSP 1629
Query: 473 IQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHLAS 532
S + QS+ SP S P L S P +P SP S + +S +
Sbjct: 1630 ---------SYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSP 1782
Query: 533 PEPNLS 538
P S
Sbjct: 1783 SSPTYS 1800
Score = 32.7 bits (73), Expect = 0.60
Identities = 31/112 (27%), Positives = 44/112 (38%), Gaps = 1/112 (0%)
Frame = +1
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPY 361
S +P +P Q A + S +SP S PS +P + +PTS S PTS P
Sbjct: 1609 SYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPT-SPNYSPTSPSYSPTSPSYSPS 1785
Query: 362 DPLSSEPIVQDQPEPNQTEPQPRTSDPS-APRASERPAARTTDTDSSTAFTP 412
P S P + P S P +P P+ S++ +TP
Sbjct: 1786 SPTYS----PSSPYNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYTP 1929
>TC226995 similar to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (59%)
Length = 679
Score = 43.1 bits (100), Expect = 4e-04
Identities = 41/133 (30%), Positives = 55/133 (40%), Gaps = 18/133 (13%)
Frame = +3
Query: 302 SSNPLTPIPE--------SQPAAQTTSPPHSPRSSFFQPSPTEAPL--------WNLLQN 345
S+ P++P P S P TTSPP SP S SPT P ++
Sbjct: 213 STTPISPTPPPSAAGSTPSSPTRSTTSPP-SPTSPTHSRSPTTPPTSSPPAPSAYSRPIA 389
Query: 346 PTS-RSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASER-PAARTTD 403
PTS + PTS T P P S P ++ +P P P T +AP AR T
Sbjct: 390 PTSPTTAAPTSATTKPAPPRCSAPPLRRNRKPLACTPAPHTPAANAPTTGTP*TTARPTP 569
Query: 404 TDSSTAFTPVSFP 416
+ +TA + + P
Sbjct: 570 SSPATASSLTNDP 608
>TC219334 weakly similar to UP|Q9FUZ9 (Q9FUZ9) SKP1 interacting partner 6
(Fragment), partial (29%)
Length = 1451
Score = 43.1 bits (100), Expect = 4e-04
Identities = 39/130 (30%), Positives = 58/130 (44%), Gaps = 5/130 (3%)
Frame = +1
Query: 304 NPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNL-LQNPTSRSEDPTSLLTIPYD 362
+P +P P + + +S PH P S+ PS T L + +SRS T L P
Sbjct: 133 SP*SPNPSTPSSLPQSSSPHEPSSNAPNPSSTSLSTHTTPLSSNSSRST--TQTLITP-- 300
Query: 363 PLSSEPIVQDQPEPNQTEPQPRTSDPS----APRASERPAARTTDTDSSTAFTPVSFPIN 418
SS P + P P+ P P ++ PS AP + P + T +STA + V +
Sbjct: 301 --SSLPFPRSPPPPS-APPTPSSAPPSTSSAAPSTTSPPPTSGSSTAASTAGSAVPPCAS 471
Query: 419 VIDSPPSNTS 428
+SPP +S
Sbjct: 472 AANSPPRESS 501
Score = 29.3 bits (64), Expect = 6.6
Identities = 18/62 (29%), Positives = 28/62 (45%)
Frame = +1
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDP 353
P+S+ S+ P S P ++ PP +P P+P+ AP P++ S P
Sbjct: 250 PLSSNSSRSTTQTLITPSSLPFPRSPPPPSAP------PTPSSAPPSTSSAAPSTTSPPP 411
Query: 354 TS 355
TS
Sbjct: 412 TS 417
>TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {Arabidopsis
thaliana;} , partial (68%)
Length = 1307
Score = 43.1 bits (100), Expect = 4e-04
Identities = 42/130 (32%), Positives = 59/130 (45%), Gaps = 2/130 (1%)
Frame = +2
Query: 304 NPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDP 363
NP TP S P +T+ S R +F P+ PL + + PT PT+ P P
Sbjct: 518 NPSTPPKPSAPPTTSTASSASLRLAF----PSAPPLVSAMP-PT-----PTNAPPPPSPP 667
Query: 364 LSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSP 423
S+ P P P+ + S PS P ++ A RT+ + SS+ P S P + +P
Sbjct: 668 PSATPPSSTSPSPSTSSTSAAPSPPSTPSSNTPSARRTSSSTSSSP-KPTSNPSS---NP 835
Query: 424 PSN--TSESI 431
PS TS SI
Sbjct: 836 PSLN*TSRSI 865
Score = 42.7 bits (99), Expect = 6e-04
Identities = 37/129 (28%), Positives = 52/129 (39%), Gaps = 2/129 (1%)
Frame = +2
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPY 361
SS P P S P++ T S + S+ P PT P NP S + S+ IP
Sbjct: 713 SSTSAAPSPPSTPSSNTPSARRTSSSTSSSPKPTSNPS----SNPPSLN*TSRSITLIPR 880
Query: 362 DPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVID 421
+ P D+P N + TS S+ AS T SST P+ V++
Sbjct: 881 SSAT*SPPPSDKPSNNPSTTPEITSLTSSNPASRGLYTSTPIWFSSTTLPSSGAPV*VLE 1060
Query: 422 --SPPSNTS 428
+PPS +
Sbjct: 1061PLAPPSTAT 1087
>TC223461 similar to UP|C941_VICSA (O81117) Cytochrome P450 94A1
(P450-dependent fatty acid omega-hydroxylase) , partial
(35%)
Length = 720
Score = 43.1 bits (100), Expect = 4e-04
Identities = 53/184 (28%), Positives = 72/184 (38%), Gaps = 5/184 (2%)
Frame = -3
Query: 305 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPL 364
PL P+PE +A + PP S SS P P+ W+ + P + T P+ P
Sbjct: 568 PL*PLPEPP*SASESQPPTS--SSA*MPFPSPNKSWDQPETPDK*TNPFPQTSTFPFQP- 398
Query: 365 SSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPIN----VI 420
S I PN P+P+ S PS+P + T DT ++ +P S P ++
Sbjct: 397 -SPTIFLA*TPPN---PRPKYS-PSSP-------SHTRDTSPTSLSSPPSHPAKRRRPLL 254
Query: 421 DSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQ-PDPEP 479
S PS E R CPS R PE LV P P + P P P
Sbjct: 253 PSSPSTP--------EGRASCSHTTLHGCKPCPSLHR----TPESLVSPLSPSKPPSPRP 110
Query: 480 AQSV 483
S+
Sbjct: 109 GSSI 98
>TC207334 similar to UP|Q949K3 (Q949K3) Suppressor-like protein, partial
(53%)
Length = 1828
Score = 42.7 bits (99), Expect = 6e-04
Identities = 42/134 (31%), Positives = 57/134 (42%), Gaps = 13/134 (9%)
Frame = -2
Query: 310 PESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIP--------- 360
P S +P HS S Q +P EA L + +P S P S L++P
Sbjct: 549 PWSAEKCSYNTPEHSASISGTQRNP-EAVLGSTHCSPNSS*TPPKSSLSLPPRLASAADT 373
Query: 361 ---YDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVS-FP 416
LSS P Q P+++EP + S S P S P ART+ + SS TP F
Sbjct: 372 SSPTHTLSSSPAPPPQTLPSRSEPASQGS-TSEPETSAAPTARTSPSTSSPKHTPTPLFS 196
Query: 417 INVIDSPPSNTSES 430
++ SP S + S
Sbjct: 195 LSPQTSPSSPRTSS 154
>AW348715
Length = 713
Score = 42.7 bits (99), Expect = 6e-04
Identities = 37/116 (31%), Positives = 48/116 (40%), Gaps = 11/116 (9%)
Frame = +2
Query: 300 QHSSNPLT------PIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNP--TSRSE 351
Q SS P T P P P + SPP P S +P + P + L +P + SE
Sbjct: 227 QSSSQPPTCTASSPPAPPRTPVPRAASPPTHPLCSCTSAAPGQTPPKSTLLSPPISPTSE 406
Query: 352 DPTSLLTIPYDPLSSEPIVQDQP-EPNQTEPQPRTSDPSAPRASER--PAARTTDT 404
DP P+ P P + P EP P+ RT P A +S + PA T T
Sbjct: 407 DPPQ----PHAPEKCTPSTEPDPSEPPTVAPRSRTRQPPATASSPQLAPAPCTLPT 562
>TC208362 weakly similar to UP|O64835 (O64835) Expressed protein
(At2g23670/F26B6.32), partial (72%)
Length = 805
Score = 42.7 bits (99), Expect = 6e-04
Identities = 35/108 (32%), Positives = 42/108 (38%)
Frame = -2
Query: 305 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPL 364
PL P P Q + PP SP PSP P+ S P+ IPY P
Sbjct: 441 PLPPPLPPAPTPQNSPPPQSPSPLPLSPSPPP-------PTPSPPSPAPSPPPPIPY-PQ 286
Query: 365 SSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTP 412
P+ Q P P P +SD + R E PA RT ST +P
Sbjct: 285 PQIPLSQSPP-----LPPPPSSDSAVSRTRECPAPRTPLPSPSTPSSP 157
>NP005385 DNA-directed RNA polymerase
Length = 2934
Score = 42.7 bits (99), Expect = 6e-04
Identities = 56/220 (25%), Positives = 74/220 (33%), Gaps = 6/220 (2%)
Frame = +1
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDP 353
P S SS +P + PA TSP +SP S + P+ + +PTS S P
Sbjct: 2281 PTSPTYSPSSPGYSP---TSPAYSPTSPSYSPTSPAYSPTSPAYSSTSPAYSPTSPSYSP 2451
Query: 354 TSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPV 413
TS P P S P P + P++P S + + + S + +P
Sbjct: 2452 TSPAYSPTSPSYSLTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPSYSPTSPSYSPTSPS 2631
Query: 414 SFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCP-----SPRRYPGPRPERLVD 468
P + SP S S + Y T P SP P V
Sbjct: 2632 YNPQSAKYSPSLAYSPSSPRLSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVS 2811
Query: 469 PD-EPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSD 507
PD P P P+ S SP TS TSD
Sbjct: 2812 PDYSPTSPQFSPSAGYSPSQPGYSPS---STSQDTPQTSD 2922
Score = 38.9 bits (89), Expect = 0.008
Identities = 61/259 (23%), Positives = 88/259 (33%), Gaps = 3/259 (1%)
Frame = +1
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDP 353
P L S +P + ++SP +SP S + PS +PTS P
Sbjct: 2104 PNLRLSPSSDAQFSPYVGGMAFSPSSSPGYSPSSPGYSPSSPG-------YSPTSPGYSP 2262
Query: 354 TSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPV 413
TS P P S P P + P++P S A ++ + + + +P
Sbjct: 2263 TSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPAYSPTSPAYSSTSPAYSPTSPS 2442
Query: 414 SFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPI 473
P + SP S + S Y P+ Y P P
Sbjct: 2443 YSPTSPAYSPTSPSYSLTSPSYSPTSPSYSPTSPSY--SPTSPAYSPTSPSY-----SPT 2601
Query: 474 QPDPEP-AQSVSNQSSVRSPHPLVETSDPHLGTSDPN-APMINIGSPQGASEAHSSNHLA 531
P P + S + QS+ SP S P L + PN +P SP S + SS +
Sbjct: 2602 SPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPTSPNYSPTSPSYSPTSPSYSPSSPTYS 2781
Query: 532 SPEP-NLSIIPYTHLQPTS 549
P N + P PTS
Sbjct: 2782 PSSPYNSGVSP--DYSPTS 2832
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.128 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,786,564
Number of Sequences: 63676
Number of extensions: 546008
Number of successful extensions: 7396
Number of sequences better than 10.0: 525
Number of HSP's better than 10.0 without gapping: 5084
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6367
length of query: 742
length of database: 12,639,632
effective HSP length: 104
effective length of query: 638
effective length of database: 6,017,328
effective search space: 3839055264
effective search space used: 3839055264
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0359.4


