

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0359.12
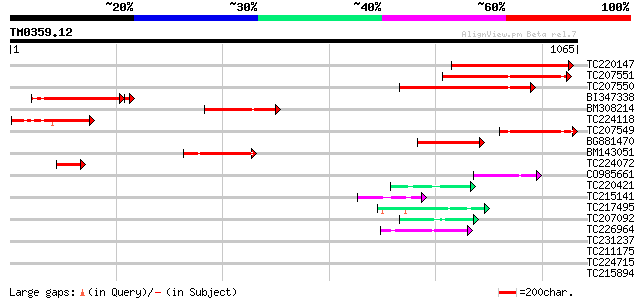
(1065 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220147 372 e-103
TC207551 289 5e-78
TC207550 262 5e-70
BI347338 238 7e-64
BM308214 187 3e-47
TC224118 similar to UP|Q940Z3 (Q940Z3) At2g36480/F1O11.11 (Expre... 185 1e-46
TC207549 168 1e-41
BG881470 135 1e-31
BM143051 128 1e-29
TC224072 similar to UP|Q940Z3 (Q940Z3) At2g36480/F1O11.11 (Expre... 86 7e-17
CO985661 72 1e-12
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 46 1e-04
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 45 1e-04
TC217495 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, part... 45 2e-04
TC207092 similar to UP|Q94ET8 (Q94ET8) LCI5, partial (11%) 44 3e-04
TC226964 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 43 6e-04
TC231237 weakly similar to GB|AAL69460.1|18491271|AY074644 At2g4... 42 0.001
TC211175 weakly similar to UP|Q9LIE8 (Q9LIE8) Similarity to cell... 41 0.003
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 41 0.003
TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase ... 40 0.004
>TC220147
Length = 1630
Score = 372 bits (956), Expect = e-103
Identities = 181/230 (78%), Positives = 199/230 (85%), Gaps = 1/230 (0%)
Frame = +3
Query: 830 EDQSDSITTSSSLPGASVSGSA-VPVPSTKDEVDDGAKTPISLSESTSTEIRNFIGFEFK 888
E+QSDSITT+SSLP ASVSGSA VPVPSTKDEVDD A+TPISLSESTS IRN IG EFK
Sbjct: 3 EEQSDSITTTSSLPVASVSGSATVPVPSTKDEVDDTARTPISLSESTSPGIRNLIGLEFK 182
Query: 889 PDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHATREREHSGLIKASRWY 948
PDVIRE H SV+SGLFD+FPH CSIC KL+FQEQF+RHL+WHATRE E +GLI ASRWY
Sbjct: 183 PDVIREFHSSVVSGLFDNFPHQCSICGHKLRFQEQFNRHLKWHATRESEENGLISASRWY 362
Query: 949 LKSSDWVVGKVESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYC 1008
LKS+DW++GK E SENEF DSVD YGKE D++QEDAMVLADE QCLCVLCGELFE+FYC
Sbjct: 363 LKSNDWILGKAEYPSENEFTDSVDTYGKEADKSQEDAMVLADEKQCLCVLCGELFEDFYC 542
Query: 1009 QENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSENPVSSVL 1058
QE GEWMFKGAVY+ NSD +MGIRD STG GPIIHA CLS+N VSSVL
Sbjct: 543 QETGEWMFKGAVYLANSDSKSEMGIRDVSTGRGPIIHASCLSDNSVSSVL 692
>TC207551
Length = 911
Score = 289 bits (739), Expect = 5e-78
Identities = 149/243 (61%), Positives = 179/243 (73%), Gaps = 2/243 (0%)
Frame = +3
Query: 814 ETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSA-VPVPSTKDEVDDGAKTPISLS 872
ETE+ VPS +DQ++ ITTS SLP S+SGSA VPV S+ DEVD KT ++
Sbjct: 3 ETESPTTVPSVAPKGSKDQTEIITTSCSLPVTSISGSAAVPVSSSDDEVDAATKTCLASP 182
Query: 873 ESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHA 932
+STSTEIRN IGF+F+P+VIRE HPSVI L+DD PHHC +C +KLK +E F+RHLEWHA
Sbjct: 183 QSTSTEIRNLIGFDFRPNVIREFHPSVIRELWDDIPHHCKVCGIKLKQEELFNRHLEWHA 362
Query: 933 TREREHSGLIKASR-WYLKSSDWVVGKVESLSENEFADSVDRYGKETDRNQEDAMVLADE 991
TRE G IKASR WY +SSDW+ GK E SE+ F DSVD + ++TD +Q D MVLADE
Sbjct: 363 TREH---GPIKASRSWYAESSDWIAGKAEYSSESGFNDSVDVHEQKTDSSQXDTMVLADE 533
Query: 992 NQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSE 1051
NQCLCVLCGELFE+ YC E EWMFKGAVY+ SD+N +M R+ GPIIHA+CLSE
Sbjct: 534 NQCLCVLCGELFEDAYCHERNEWMFKGAVYMNYSDVNCEMESRNV----GPIIHAKCLSE 701
Query: 1052 NPV 1054
N V
Sbjct: 702 NSV 710
>TC207550
Length = 775
Score = 262 bits (670), Expect = 5e-70
Identities = 149/259 (57%), Positives = 183/259 (70%), Gaps = 4/259 (1%)
Frame = +1
Query: 733 LIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPP-RTSALPPASSNLKSAAVQASSSA 791
LI SA+ASP SL ++SS LPK P +A QPP R S PPASS++ S++ ++A
Sbjct: 10 LISSGSAVASPSSLDPLHNDSSTLPKKPQGKAGQPPQRLSTQPPASSSVSSSSAPTLNAA 189
Query: 792 -NNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGS 850
NN LNPIANLL+SLVAKGLISAETE+ VPSE +DQ++ ITTS SLP S+SGS
Sbjct: 190 KNNKLNPIANLLSSLVAKGLISAETESPTTVPSEAPKGSKDQTEIITTSCSLPVTSISGS 369
Query: 851 A-VPVPSTKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPH 909
A +PV S+ D+VD K + +STSTEIRN IGF+F+P+VIRE HPSVI L+DDFPH
Sbjct: 370 AAIPVSSSGDKVDAATKISHASPQSTSTEIRNLIGFDFRPNVIREFHPSVIRELWDDFPH 549
Query: 910 HCSICSLKLKFQEQFDRHLEWHATREREHSGLIKASR-WYLKSSDWVVGKVESLSENEFA 968
+C +C +KLK QE F+RHLEWHA RE G IKASR WY KS DW+ G+ E SE+EF
Sbjct: 550 NCKVCGIKLK-QELFNRHLEWHAAREH---GPIKASRSWYAKSIDWIAGRTEYSSESEFT 717
Query: 969 DSVDRYGKETDRNQEDAMV 987
DSVD K+ D +Q D MV
Sbjct: 718 DSVDLQDKKIDSSQLDTMV 774
>BI347338
Length = 581
Score = 238 bits (607), Expect(2) = 7e-64
Identities = 125/175 (71%), Positives = 139/175 (79%)
Frame = +2
Query: 41 LPQRQQQVAASGISTLSSAARLRAAANSRDLESSDFGRGDGGGGYHPRPPPFQELVAQYK 100
LPQR + ++TL S R A RD E SD GRG GGGGY P+PPP QELV QYK
Sbjct: 26 LPQRP-----TAVTTLPST---RFRAYGRDSEISDLGRG-GGGGYQPQPPPHQELVTQYK 178
Query: 101 SALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSIVKN 160
+ALAELTFNSKPIITNLTIIAGEN +A +AIAA V NI+EVPS+QKLPSLYLLDSIVKN
Sbjct: 179 TALAELTFNSKPIITNLTIIAGENLSAAKAIAAAVYDNILEVPSDQKLPSLYLLDSIVKN 358
Query: 161 IGRGYVKYFAARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKEL 215
IGR Y+KYFA RLPEVFCK Y+ VDP VH SM+HLFGTW+GVFP Q+LQ IEKEL
Sbjct: 359 IGRDYIKYFAYRLPEVFCKTYKQVDPCVHSSMQHLFGTWKGVFPPQSLQMIEKEL 523
Score = 25.8 bits (55), Expect(2) = 7e-64
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = +1
Query: 217 FTPAVNGSSSTSATLRSD 234
F PAVN S+S SAT R D
Sbjct: 526 FAPAVNSSASVSATDRRD 579
>BM308214
Length = 428
Score = 187 bits (474), Expect = 3e-47
Identities = 98/144 (68%), Positives = 117/144 (81%), Gaps = 2/144 (1%)
Frame = +1
Query: 367 GQRNGLSIKNNFLNTEA--SMILDARNQPRQNINSLQRGVMSNSWKHSEEEEFMWDEMNA 424
GQ NGL +K +FLNTEA SMILD +QP QNI+S + V+S SWK+SEEEE+ WDEMN+
Sbjct: 1 GQGNGLGLKYSFLNTEAPKSMILDVHHQPTQNISSTRTSVISASWKNSEEEEYTWDEMNS 180
Query: 425 GLTGHDAASVPNNLSKDSWTSDDENLEVEDNHHQSRNPFGANVDTEMSIESQATERKQLP 484
GLT H A++V +NLSK+SWT+DDENLE ED + RNPF ANVD EMSIESQATE+KQLP
Sbjct: 181 GLTVHGASTV-SNLSKNSWTADDENLEAEDCL-EIRNPFRANVDREMSIESQATEKKQLP 354
Query: 485 AFRPHPSLSWKLQEQHSIDELNQK 508
+ HPSLSW+LQEQ SIDELN+K
Sbjct: 355 SSHHHPSLSWQLQEQQSIDELNRK 426
>TC224118 similar to UP|Q940Z3 (Q940Z3) At2g36480/F1O11.11 (Expressed
protein), partial (46%)
Length = 891
Score = 185 bits (469), Expect = 1e-46
Identities = 106/159 (66%), Positives = 119/159 (74%), Gaps = 4/159 (2%)
Frame = +1
Query: 4 SSRRPFDRSREPGLKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAARLR 63
+SRRPFDRSREPG K+PR+MEE +R A N +RQF Q+ ASGI+TLSSA R
Sbjct: 406 NSRRPFDRSREPGPKKPRLMEELDR---ASNSGARQF----HQRQVASGITTLSSA---R 555
Query: 64 AAANSRDLESSDFGR----GDGGGGYHPRPPPFQELVAQYKSALAELTFNSKPIITNLTI 119
N RDLESSDFGR G GGGGY P+P PFQELVAQYK+ALAELTFNSKPIITNLTI
Sbjct: 556 FRTNDRDLESSDFGRRGGAGGGGGGYQPQPLPFQELVAQYKAALAELTFNSKPIITNLTI 735
Query: 120 IAGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSIV 158
IAGENQAAE+AIAATVCANI+EV L+L +V
Sbjct: 736 IAGENQAAEKAIAATVCANILEVNLVYSTVQLFLCSIVV 852
>TC207549
Length = 812
Score = 168 bits (425), Expect = 1e-41
Identities = 89/146 (60%), Positives = 103/146 (69%), Gaps = 1/146 (0%)
Frame = +3
Query: 921 QEQFDRHLEWHATREREHSGLIKASR-WYLKSSDWVVGKVESLSENEFADSVDRYGKETD 979
QE F+RHLEWHA RE G IKASR WY KS DW+ G+ E SE+EF DSVD K+ D
Sbjct: 9 QELFNRHLEWHAAREH---GPIKASRSWYAKSIDWIAGRTEYSSESEFTDSVDLQDKKID 179
Query: 980 RNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTG 1039
+Q D MVLADENQCLCVLCGELFE+ C + EWMFKGAVY+ SD+N +M R+
Sbjct: 180 SSQLDTMVLADENQCLCVLCGELFEDVCCHDRNEWMFKGAVYMNFSDVNCEMESRNV--- 350
Query: 1040 GGPIIHARCLSENPVSSVLNMVRLII 1065
GPIIHA+CLSEN V + N VR II
Sbjct: 351 -GPIIHAKCLSENSV--ITNSVRPII 419
>BG881470
Length = 382
Score = 135 bits (340), Expect = 1e-31
Identities = 74/127 (58%), Positives = 95/127 (74%), Gaps = 2/127 (1%)
Frame = +2
Query: 767 PPRTSALPPASSNLKSAAVQASSSA-NNALNPIANLLNSLVAKGLISAETETTAKVPSEM 825
P R S PPASSN+ S++ ++A NN LNPI+NLL+SLVAKGLISAETE+ VPSE+
Sbjct: 2 PQRLSTQPPASSNISSSSAPTLNTAKNNKLNPISNLLSSLVAKGLISAETESPTMVPSEV 181
Query: 826 LTRLEDQSDSITTSSSLPGASVSGS-AVPVPSTKDEVDDGAKTPISLSESTSTEIRNFIG 884
+DQ++ ITTS SLP S+SGS AVPV S+ DEVD KT ++ +STSTEIRN +G
Sbjct: 182 PKGSKDQTEIITTSCSLPVTSISGSAAVPVSSSGDEVDSATKTSLASPQSTSTEIRNLVG 361
Query: 885 FEFKPDV 891
F+F+P+V
Sbjct: 362 FDFRPNV 382
>BM143051
Length = 440
Score = 128 bits (322), Expect = 1e-29
Identities = 70/140 (50%), Positives = 90/140 (64%), Gaps = 2/140 (1%)
Frame = +2
Query: 326 SDLSRNLGMGTGRTGGRVAELGSDKSGYNKAAGAGVAGTISGQRNGLSIKNNFLNTEASM 385
S +S NL G GRTG ++ +LG DK+ + G T SGQRNG ++K ++ N EA
Sbjct: 32 SVISSNLVSGAGRTGSKLIDLGHDKTWFK--TDGGDPDTTSGQRNGFNLKRSYSNREAPK 205
Query: 386 I--LDARNQPRQNINSLQRGVMSNSWKHSEEEEFMWDEMNAGLTGHDAASVPNNLSKDSW 443
+ LDA QPRQ+ ++ +MS +WK SEEEEFMW EMN GLT H A+V +NLS D+W
Sbjct: 206 LTNLDAHRQPRQSTTDIRNNLMSGNWKTSEEEEFMWGEMNIGLTDH-GANVSSNLSTDTW 382
Query: 444 TSDDENLEVEDNHHQSRNPF 463
+DDENLE ED H Q PF
Sbjct: 383 MADDENLEGED-HLQITRPF 439
>TC224072 similar to UP|Q940Z3 (Q940Z3) At2g36480/F1O11.11 (Expressed
protein), partial (37%)
Length = 393
Score = 86.3 bits (212), Expect = 7e-17
Identities = 44/55 (80%), Positives = 48/55 (87%)
Frame = +3
Query: 88 RPPPFQELVAQYKSALAELTFNSKPIITNLTIIAGENQAAEQAIAATVCANIIEV 142
RPPP QELV QYK+ALAELTFNSKPIITNLTIIAGEN +A +AIAA V NI+EV
Sbjct: 6 RPPPHQELVTQYKTALAELTFNSKPIITNLTIIAGENLSAAKAIAAAVYDNILEV 170
>CO985661
Length = 553
Score = 72.0 bits (175), Expect = 1e-12
Identities = 38/130 (29%), Positives = 66/130 (50%), Gaps = 2/130 (1%)
Frame = -2
Query: 871 LSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEW 930
+S + ++ +G EF PD+++ H S ++ L+ D P C+ C+ + K QE+ H++W
Sbjct: 459 ISLANQLPAQDXVGTEFNPDILKIRHESAVNALYGDLPRQCTSCAXRFKCQEEHSSHMDW 280
Query: 931 HATRER-EHSGLIKASR-WYLKSSDWVVGKVESLSENEFADSVDRYGKETDRNQEDAMVL 988
H T+ R S K SR W++ W+ G E+L + E ++ E+ V
Sbjct: 279 HVTKNRMSKSRKQKPSRKWFVSDRMWLSG-AEALGTESAPGFLPTETIEEMKDHEELAVP 103
Query: 989 ADENQCLCVL 998
A+E+Q C L
Sbjct: 102 AEEDQNTCAL 73
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 45.8 bits (107), Expect = 1e-04
Identities = 44/162 (27%), Positives = 64/162 (39%), Gaps = 1/162 (0%)
Frame = +3
Query: 715 PSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALP 774
PS PS +S S + PP S SPPS P P P + P SA P
Sbjct: 261 PSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSP-------PSAPSPSSTAPSPPSASP 419
Query: 775 PASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSD 834
P S + + +++SAN+ A G ++ T T+ P+ S
Sbjct: 420 PTSPSPSPPSPASTASANSP------------ASGSPTSRTSPTSPSPTSTSKPPAPSSS 563
Query: 835 SITTSSSLPGASVSG-SAVPVPSTKDEVDDGAKTPISLSEST 875
S SS P S + S P P+T + +PI+ S++T
Sbjct: 564 SPA*PSSKPSPSPTPISPDPSPATSTPTSHTSISPITASKAT 689
Score = 40.8 bits (94), Expect = 0.003
Identities = 39/141 (27%), Positives = 54/141 (37%), Gaps = 3/141 (2%)
Frame = +3
Query: 662 TEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVR 721
T PH + P P T P+ T +A S PTS S + PS
Sbjct: 210 TSPPHRTPAPSPPSTMPPSATPPSPSATSSPSAS----PTAPPTSPSPSPPSSPSPPSAP 377
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRR---AAQPPRTSALPPASS 778
S+ SP PP S SPPS +S P R + P +++ PPA S
Sbjct: 378 SPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPPAPS 557
Query: 779 NLKSAAVQASSSANNALNPIA 799
+ S+ SS + + PI+
Sbjct: 558 S--SSPA*PSSKPSPSPTPIS 614
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 45.4 bits (106), Expect = 1e-04
Identities = 36/132 (27%), Positives = 54/132 (40%), Gaps = 2/132 (1%)
Frame = +2
Query: 653 SSHTEVTAKTEK--PHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLD 710
SS T + T PH P T PT+ A+ +L N P N
Sbjct: 515 SSSTSSPSPTSSTWPHRPASATPCRTPNPTSPPTSLASSTFSRLPNPPTPN--------- 667
Query: 711 TGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRT 770
P + G+RP+ S+ S+P L P +A SPP+ S+ P+ P R++ P T
Sbjct: 668 ----PPLSGLRPAPSTASTPKTLSPSSTAPTSPPA-------STPPPRRPARKSPTPTTT 814
Query: 771 SALPPASSNLKS 782
P+ ++ S
Sbjct: 815 FTASPSPDSVSS 850
Score = 34.3 bits (77), Expect = 0.30
Identities = 50/197 (25%), Positives = 78/197 (39%), Gaps = 19/197 (9%)
Frame = +2
Query: 704 PTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPP------SLGRPKDNSSALP 757
P+S + + + L S SS SSP+ +P + PP S P +S P
Sbjct: 89 PSSSTPPPNSSSAPLSWSHSSSSSSSPSTTLP-----SPPPTPPTATSTPTPTTSSPPPP 253
Query: 758 KIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETET 817
P R + P A P AS +L S ++AS +A P + V+ GL ++ T T
Sbjct: 254 SAPGRSRSATPPPPAAPTASPSL-SLGLRASWAAT---APWP*RSAATVSWGLTTSTTTT 421
Query: 818 T--AKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVP-----------STKDEVDDG 864
T P + S TS++ P + S ++ P P +T +
Sbjct: 422 TLPLNAPGRQCSGNTKCSSWKVTSTTPPCSRSSSTSSPSPTSSTWPHRPASATPCRTPNP 601
Query: 865 AKTPISLSESTSTEIRN 881
P SL+ ST + + N
Sbjct: 602 TSPPTSLASSTFSRLPN 652
>TC217495 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, partial (59%)
Length = 2299
Score = 44.7 bits (104), Expect = 2e-04
Identities = 59/233 (25%), Positives = 91/233 (38%), Gaps = 22/233 (9%)
Frame = +2
Query: 691 GGKLSNMPI--------TNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIAS 742
GG+L+ P+ N LP P L + S + PP+ ++S
Sbjct: 602 GGRLAMPPMYWQGYYGAPNGLPQLHQQSLLQPPPGLSMPSSMQQPMQYPNITPPLPTVSS 781
Query: 743 ----------PPSLGRPKDNSSALP--KIPPRRAAQPPRTSALPPASSNLKSAAVQASSS 790
P S P S++LP +PP +A P SALPPA S L S + A S
Sbjct: 782 NLPELPSSLLPASASTPSITSASLPPSNLPPAPSALAPAPSALPPAPSAL-SPSSSALSP 958
Query: 791 ANNALNPIANLLNSLV--AKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVS 848
A +A NL S+ A + ++ A +PS LT ++I S ++S
Sbjct: 959 APSATLASENLPVSVTNKAPNVSTSAAMLAANLPS--LTISGPDINAIVPPISSKPHAIS 1132
Query: 849 GSAVPVPSTKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVIS 901
GS++P + +P + STS P + + PS++S
Sbjct: 1133GSSLPYQTVSQ------FSPAVVGSSTSIHTETSAPSLLTPGQLLQPGPSIVS 1273
>TC207092 similar to UP|Q94ET8 (Q94ET8) LCI5, partial (11%)
Length = 1015
Score = 44.3 bits (103), Expect = 3e-04
Identities = 35/148 (23%), Positives = 59/148 (39%)
Frame = +1
Query: 733 LIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSAN 792
L P S+ SPP+ P ++A PPR ++ + L ++ + +A+ S S
Sbjct: 124 LTPKTSSFCSPPTASVPTAVAAAASSTPPRSSSSSSSPAPLSSSTPPTRRSALPESDSTA 303
Query: 793 NALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAV 852
+ P +G S T+ PS + + +E S S T S P A+ + S+
Sbjct: 304 SGSEP---------TRGPFS-----TSPSPSPLKSTIETSSPSPMTPSPSPSATAAASSA 441
Query: 853 PVPSTKDEVDDGAKTPISLSESTSTEIR 880
P+ P S S T++R
Sbjct: 442 SSPAAAAAASGRVAPPTSTPRSPLTDLR 525
>TC226964 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (5%)
Length = 1717
Score = 43.1 bits (100), Expect = 6e-04
Identities = 45/174 (25%), Positives = 77/174 (43%), Gaps = 1/174 (0%)
Frame = +3
Query: 696 NMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSA 755
+MP + + P T LP++ S + P+ L+P ++ S S P + A
Sbjct: 90 SMPSSMQQPMQYPNFTPPLPTV-----SSNLPELPSSLLPVSASTPSVTSASLPPNLPPA 254
Query: 756 LPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAET 815
+PP +A PP SAL P S ++ + S AN A P + +++A L S
Sbjct: 255 PSALPPAPSALPPAPSALSPVPSATLASEIFPVSVANKA--PNVSTSAAMLASNLPSLAL 428
Query: 816 ETT-AKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTP 868
T A+ + ++ + +S++I + SSLP SVS + V + + P
Sbjct: 429 LTNPARDINAIVPPISSKSNAI-SGSSLPYQSVSQLSPAVVESSTSIHTETSAP 587
>TC231237 weakly similar to GB|AAL69460.1|18491271|AY074644
At2g41330/F13H10.12 {Arabidopsis thaliana;} , partial
(24%)
Length = 583
Score = 42.4 bits (98), Expect = 0.001
Identities = 52/206 (25%), Positives = 80/206 (38%), Gaps = 10/206 (4%)
Frame = +2
Query: 685 ETAAGKGGKLSNMPITN-RLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPP------- 736
+T+ K N P+ N RL ++ S G P S+ +P PP
Sbjct: 2 KTSRASSKKNPNRPLRNLRLSSAAS----------GSPPPCSAPGAPHAPPPPPHSRPAS 151
Query: 737 --VSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNA 794
VS+ SPPS +++A P P A+ P TSA P S SA SSA A
Sbjct: 152 TRVSSSTSPPSASSAAPSTTAAPSDPSSAASASPSTSATSP--STTASATSSTPSSAAAA 325
Query: 795 LNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPV 854
+P +S+ +T+ P+ + S +SS ++ S +
Sbjct: 326 TSPCR-----------VSSSAASTSAAPT-------TSASSTRAASSTASSNASRAPT*T 451
Query: 855 PSTKDEVDDGAKTPISLSESTSTEIR 880
P+T E D + +S E+T +R
Sbjct: 452 PATPAEASD-SLCAMSAMEATKCSLR 526
>TC211175 weakly similar to UP|Q9LIE8 (Q9LIE8) Similarity to cell wall-plasma
membrane linker protein, partial (3%)
Length = 609
Score = 40.8 bits (94), Expect = 0.003
Identities = 40/138 (28%), Positives = 61/138 (43%), Gaps = 2/138 (1%)
Frame = +1
Query: 722 PSQSSGSSPAGLIPPVSAI-ASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNL 780
PS S SP PP A +PP+ S P + P + PP T+ PPA
Sbjct: 154 PSPKSSPSPPTAKPPSPATHPAPPAKSPTLSPPSQSPAVSPSGSTPPPATN--PPA---- 315
Query: 781 KSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSS 840
KS A Q SS A +P N+ + + T A V S + ++ + +S+
Sbjct: 316 KSPATQPPSSVPPASSPSNNVSSPPAPSP--ATPPPTVAPVSSPVGAPPVAEAPTPESST 489
Query: 841 SLPGASV-SGSAVPVPST 857
+P ++V +GS + +PST
Sbjct: 490 GIPSSAVPAGSPITLPST 543
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 40.8 bits (94), Expect = 0.003
Identities = 28/83 (33%), Positives = 43/83 (51%)
Frame = +2
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLK 781
P+ + SSP +PP+ A + PPS P + S+ P P P TS PP+ S
Sbjct: 620 PASPTTSSPTTALPPLPASSPPPS---PMHSPSSSPS--PSAPTSPAATST-PPSPSAPS 781
Query: 782 SAAVQASSSANNALNPIANLLNS 804
SAA SS+A++ +P ++ +S
Sbjct: 782 SAATSPSSAASSTSSPSSSAPSS 850
>TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase , complete
Length = 1675
Score = 40.4 bits (93), Expect = 0.004
Identities = 48/184 (26%), Positives = 72/184 (39%), Gaps = 1/184 (0%)
Frame = +1
Query: 695 SNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSS 754
S+ P T P+ R PS L P+ SS S+P +P S SS
Sbjct: 283 SSAPATWEWPSRR-------PSSLRTSPTSSSSSTP-----------TPTSSAARCSTSS 408
Query: 755 ALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAE 814
P P + PP T PPA ++ S A S A++A S +S+
Sbjct: 409 TPPPSSPAPRSTPPPTPPSPPAPTSASSPPAPARSPASHASTSSRGTSPSSAPLFPLSSV 588
Query: 815 TETTAKVPSEMLTRLEDQSDSITTS-SSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSE 873
T P+++ + S S TS S P + + S+ P P T + +PI+L+
Sbjct: 589 TP-----PTQLSSSFPTPSTSSPTSRGSSPASPPTASSAPAP-TWTLPASVSSSPITLTS 750
Query: 874 STST 877
+ T
Sbjct: 751 TPRT 762
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,899,297
Number of Sequences: 63676
Number of extensions: 669503
Number of successful extensions: 5960
Number of sequences better than 10.0: 379
Number of HSP's better than 10.0 without gapping: 4894
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5561
length of query: 1065
length of database: 12,639,632
effective HSP length: 107
effective length of query: 958
effective length of database: 5,826,300
effective search space: 5581595400
effective search space used: 5581595400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0359.12


