

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
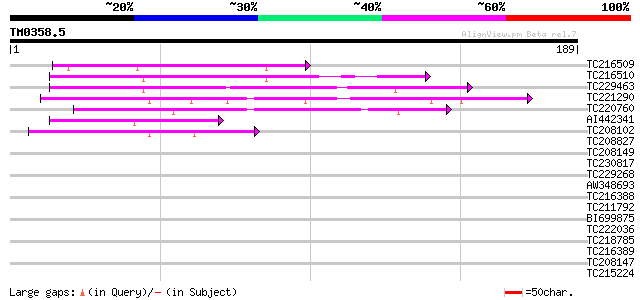
Query= TM0358.5
(189 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216509 similar to UP|O48915 (O48915) Non-race specific disease... 51 3e-07
TC216510 weakly similar to UP|Q6T3R2 (Q6T3R2) NDR1-like protein,... 51 3e-07
TC229463 weakly similar to UP|O81329 (O81329) F3D13.5 protein, p... 46 1e-05
TC221290 weakly similar to UP|Q9SHY5 (Q9SHY5) F1E22.7, partial (... 42 1e-04
TC220760 similar to UP|Q756S8 (Q756S8) AER176Wp, partial (9%) 42 2e-04
AI442341 42 2e-04
TC208102 weakly similar to UP|Q6RY60 (Q6RY60) Hin1-like protein ... 41 3e-04
TC208827 UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein B13-1-9,... 39 0.001
TC208149 homologue to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced pro... 39 0.001
TC230817 similar to UP|Q6MWW7 (Q6MWW7) PE-PGRS FAMILY PROTEIN (F... 37 0.004
TC229268 similar to GB|BAB32889.1|13122296|AB047812 hin1 homolog... 37 0.004
AW348693 weakly similar to GP|19110824|gb ABC transporter ABCA.5... 37 0.004
TC216388 similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partia... 37 0.007
TC211792 homologue to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced pro... 36 0.009
BI699875 similar to PIR|E96668|E96 protein F1N19.3 [imported] - ... 34 0.035
TC222036 similar to PIR|E96668|E96668 protein F1N19.3 [imported]... 34 0.046
TC218785 weakly similar to GB|BAB08954.1|9758412|AB006700 harpin... 33 0.079
TC216389 weakly similar to UP|Q9M663 (Q9M663) Harpin inducing pr... 33 0.079
TC208147 homologue to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced pro... 33 0.10
TC215224 32 0.13
>TC216509 similar to UP|O48915 (O48915) Non-race specific disease resistance
protein (NDR1) (At3g20600), partial (17%)
Length = 1240
Score = 51.2 bits (121), Expect = 3e-07
Identities = 34/103 (33%), Positives = 51/103 (49%), Gaps = 17/103 (16%)
Frame = +3
Query: 15 IGFL---GLLVLCLWLALRPKSPSYSVVFL-----------SIEQHPGENGSIFYSLEIE 60
IGFL GL L LWL+LR P + ++ S +N +I ++L++
Sbjct: 315 IGFLIAIGLTALFLWLSLRVDEPKCYLDYIYVPALNKTLNSSSNSTHNKNATIVFALKLA 494
Query: 61 NPNKDSSIYYDDIILSFLYGQQED---KVGETTIGSFHQGTGK 100
N NKD I YDD++LSF + + +G T+ F+QG K
Sbjct: 495 NENKDKGIQYDDVLLSFRVFESVNTTRPLGNATVERFYQGHKK 623
>TC216510 weakly similar to UP|Q6T3R2 (Q6T3R2) NDR1-like protein, partial
(35%)
Length = 1117
Score = 51.2 bits (121), Expect = 3e-07
Identities = 38/139 (27%), Positives = 62/139 (44%), Gaps = 12/139 (8%)
Frame = +1
Query: 14 FIGFLGLLVLCLWLALRPKSPSYSVVFLSI---------EQHPGENGSIFYSLEIENPNK 64
F+ +GL L LWL+LR P + ++ + +N +I ++L++ N NK
Sbjct: 310 FLITIGLTALFLWLSLRVDEPKCYLDYIYVPALNKTLNSNSTHNKNTTILFALKLTNGNK 489
Query: 65 DSSIYYDDIILSFLYGQQED---KVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKA 121
D I YDD++LSF + + +G T+ F+QG K + +HG F
Sbjct: 490 DKGIQYDDVLLSFRVFESVNLTRPLGNATVQRFYQGHQKKAT-------KHGNF------ 630
Query: 122 ISNATAELKVALTTRFQYK 140
S L A+ R Y+
Sbjct: 631 -SGGGGNLTAAVAGRMWYR 684
>TC229463 weakly similar to UP|O81329 (O81329) F3D13.5 protein, partial (66%)
Length = 1212
Score = 45.8 bits (107), Expect = 1e-05
Identities = 38/154 (24%), Positives = 72/154 (46%), Gaps = 13/154 (8%)
Frame = +3
Query: 14 FIGFLGLLVLCLWLALRPKSPSYSVVFLSI-----EQHPGENGSIFYSLEIENPNKDSSI 68
F+ G+ +L LWL RP P ++V+ +I P + ++ +S+ I+NPN+ SI
Sbjct: 141 FLLLAGVTLLVLWLVYRPHKPRFTVIGAAIYGLNTSTPPLMSTTMQFSVLIKNPNRRVSI 320
Query: 69 YYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAISNATA- 127
YYD +F+ + + + + +Q + SV + G P+ +SN A
Sbjct: 321 YYDR-FSAFVSYRNQAITPQVLLPPLYQEKRSSVSVSPVIG---GTPLPVSVEVSNGLAM 488
Query: 128 -------ELKVALTTRFQYKTWGIKSKFHGLHLQ 154
L++ R ++K IK+ +GL+++
Sbjct: 489 DEAYGVVGLRLIFQGRVRWKAGAIKTAHYGLYVK 590
>TC221290 weakly similar to UP|Q9SHY5 (Q9SHY5) F1E22.7, partial (68%)
Length = 916
Score = 42.4 bits (98), Expect = 1e-04
Identities = 50/185 (27%), Positives = 79/185 (42%), Gaps = 21/185 (11%)
Frame = +3
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ-HPGENGSIFYSLEI----ENPNKD 65
L+ I + + + L+L RPK P YSV L I Q + +N +++ + + NPNK
Sbjct: 315 LLILIIAIAITIGILYLVFRPKLPKYSVDQLRISQFNVSDNNTLYATFNVAITARNPNKK 494
Query: 66 SSIYYD-DIILSFLYGQQEDKVGETTIGSFHQG-----------TGKTRSVYDTVNARHG 113
IYY+ +S Y E ++ E ++ F+QG TG+ VN
Sbjct: 495 IGIYYEGGSHISAWY--METQLCEGSLPKFYQGHRNTTVLDLPLTGQAHDANGLVN---- 656
Query: 114 AFKPLFKAISNATAELKVALTTRFQY-KTWGIKSKFH---GLHLQGILPIDSDGKLSRKK 169
+ + +N LKV R + K K KF L + IL +D ++SR
Sbjct: 657 RIQEQLQQTNNVPLNLKVNQPVRVKLGKLKLFKVKFRVRCKLEVDNILGASNDIRISRSS 836
Query: 170 KKYPL 174
K+ L
Sbjct: 837 CKFRL 851
>TC220760 similar to UP|Q756S8 (Q756S8) AER176Wp, partial (9%)
Length = 721
Score = 42.0 bits (97), Expect = 2e-04
Identities = 32/139 (23%), Positives = 62/139 (44%), Gaps = 13/139 (9%)
Frame = +2
Query: 22 VLCLWLALRPKSPSYSVVFLSIEQHPGENGSI----FYSLEIENPNKDSSIYYDDIILSF 77
++ WL LRP P +++ LS+ + S+ S + N NK ++ Y+ + S
Sbjct: 2 LIITWLVLRPSLPHFTLHSLSVSNLSSTSQSLSATWHLSFLVRNGNKKMTVSYNALRSSI 181
Query: 78 LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAISNATAE--------- 128
Y +++ + E+ + F Q T ++ T+ A +P K I N AE
Sbjct: 182 FY--RQNYISESQLAPFRQDTRSQTTLNATLTAAGTYLEP--KLIDNLNAERNASSVLFD 349
Query: 129 LKVALTTRFQYKTWGIKSK 147
++V T F+ +W +++
Sbjct: 350 VQVVAATSFRSGSWRFRTR 406
>AI442341
Length = 410
Score = 41.6 bits (96), Expect = 2e-04
Identities = 22/63 (34%), Positives = 36/63 (56%), Gaps = 5/63 (7%)
Frame = +3
Query: 14 FIGFLGLLVLCLWLALRPKSPSYSVVF-----LSIEQHPGENGSIFYSLEIENPNKDSSI 68
F+ G+ +L LWL RP P ++V+ L+ P + ++ +S+ I+NPN+ SI
Sbjct: 69 FLLLAGVTLLVLWLVYRPHKPRFTVIGAAVYDLNTTTPPLMSTTVQFSVLIKNPNRRVSI 248
Query: 69 YYD 71
YYD
Sbjct: 249 YYD 257
>TC208102 weakly similar to UP|Q6RY60 (Q6RY60) Hin1-like protein (Fragment),
partial (25%)
Length = 823
Score = 41.2 bits (95), Expect = 3e-04
Identities = 23/83 (27%), Positives = 45/83 (53%), Gaps = 6/83 (7%)
Frame = +2
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSLEIE---- 60
F++ L+ I + L +L L++ + P+S ++++ ++ Q + N +++Y L +
Sbjct: 152 FWILLVLIITIVMLAILVLYIIITPRSFRFTLIDANLTQFDYTANNSTLYYDLVLNITAH 331
Query: 61 NPNKDSSIYYDDIILSFLYGQQE 83
NPNK IYYD + LY + E
Sbjct: 332 NPNKRLKIYYDVVRAHALYRRVE 400
>TC208827 UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein B13-1-9, complete
Length = 768
Score = 39.3 bits (90), Expect = 0.001
Identities = 22/69 (31%), Positives = 37/69 (52%), Gaps = 6/69 (8%)
Frame = +2
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSLEI----ENPNK 64
L+ I +GL+ L WL ++P+S + V + Q + N ++ Y++ + NPNK
Sbjct: 218 LVALIVLVGLVFLIFWLVVQPRSFKFQVTEADLTQFDYYTNNHTLHYNMVLNFTARNPNK 397
Query: 65 DSSIYYDDI 73
SIYYD +
Sbjct: 398 KLSIYYDKV 424
>TC208149 homologue to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein
B13-1-9, partial (59%)
Length = 422
Score = 38.9 bits (89), Expect = 0.001
Identities = 21/69 (30%), Positives = 36/69 (51%), Gaps = 6/69 (8%)
Frame = +3
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSLEIE----NPNK 64
L+ I +GL +L WL ++P+ + V + Q + N ++ Y++ + NPNK
Sbjct: 111 LVALIVLVGLAILIFWLVVQPRYFKFHVTKADLTQFDYYSNNNTLHYNMVLNFTARNPNK 290
Query: 65 DSSIYYDDI 73
SIYYD +
Sbjct: 291 KLSIYYDKV 317
>TC230817 similar to UP|Q6MWW7 (Q6MWW7) PE-PGRS FAMILY PROTEIN (Fragment),
partial (3%)
Length = 1064
Score = 37.4 bits (85), Expect = 0.004
Identities = 29/91 (31%), Positives = 42/91 (45%), Gaps = 5/91 (5%)
Frame = -3
Query: 9 VFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQH----PGE-NGSIFYSLEIENPN 63
+FL+ +G G ++ L+ RP+ P++SV L + P N +L NPN
Sbjct: 882 LFLLLLVGAAGTVLYFLY---RPQRPTFSVTSLKLSSFNLTTPSTINAKFDLTLSTTNPN 712
Query: 64 KDSSIYYDDIILSFLYGQQEDKVGETTIGSF 94
YD +S LYG + V TTI SF
Sbjct: 711 DKIIFSYDPTSVSLLYG--DTAVASTTIPSF 625
>TC229268 similar to GB|BAB32889.1|13122296|AB047812 hin1 homolog
{Arabidopsis thaliana;} , partial (31%)
Length = 1043
Score = 37.4 bits (85), Expect = 0.004
Identities = 43/160 (26%), Positives = 65/160 (39%), Gaps = 17/160 (10%)
Frame = +2
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHP-GENGSIFYSL----EIENPNKD 65
++ + LG L W +RP + V S+ + N ++ Y L I NPN+
Sbjct: 260 ILIIVAILGFL---FWFIVRPNVLKFHVTDASLTRFDYTTNNTLHYDLALNVSIRNPNRR 430
Query: 66 SSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVY-----DTVNARHGAFKPLF- 119
+YYD I LY Q+ G T+G F Q T V V G +F
Sbjct: 431 VGVYYDHIEAHALY--QDVLFGNQTLGPFFQHHKNTTFVNPLFKGQRVTPLAGNQVEVFD 604
Query: 120 --KAISNATAELKVALTTRFQ---YKTWGIKSKFH-GLHL 153
K T +LK+ + RF+ +K+ +K K LH+
Sbjct: 605 KEKGSGVYTIDLKLFMVVRFKFLLFKSASVKPKIRCALHV 724
>AW348693 weakly similar to GP|19110824|gb ABC transporter ABCA.5
{Dictyostelium discoideum}, partial (1%)
Length = 699
Score = 37.4 bits (85), Expect = 0.004
Identities = 37/152 (24%), Positives = 64/152 (41%), Gaps = 16/152 (10%)
Frame = -2
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGSIF-----YSLEIENPNKDSSIYYDD 72
+G+ V+ + L L+PK Y+V +I + + +++ NP SIYYD
Sbjct: 689 VGIAVIIIXLVLKPKRLEYTVENAAIHXXXXTDANXXXANFDFTIRSYNPXXRVSIYYDT 510
Query: 73 IILSFLYGQQEDKVGETTIGSFHQG-----------TGKTRSVYDTVNARHGAFKPLFKA 121
+ +S Y ++ + + F Q T +T ++YD+V K L
Sbjct: 509 VEVSVRY--EDQTLATNAVQPFFQSHKNVTRLHVGLTAQTVALYDSVP------KDLRLE 354
Query: 122 ISNATAELKVALTTRFQYKTWGIKSKFHGLHL 153
S+ EL V + R ++K KSK L +
Sbjct: 353 RSSGDIELDVWMRARIRFKVGVWKSKHRVLKI 258
>TC216388 similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial (43%)
Length = 1056
Score = 36.6 bits (83), Expect = 0.007
Identities = 36/151 (23%), Positives = 61/151 (39%), Gaps = 11/151 (7%)
Frame = +3
Query: 8 YVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ-HPGENGSIFYSLE----IENP 62
+ ++ I +G+ V WL +RP + V ++ Q + N ++ Y L + NP
Sbjct: 213 FKLILTVIIIIGIAVFLFWLIVRPNVVKFHVTEATLTQFNYTPNNTLHYDLALNITVRNP 392
Query: 63 NKDSSIYYDDI-ILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYD-----TVNARHGAFK 116
NK IYYD I + + + D H+ T V+ +NA A
Sbjct: 393 NKRLGIYYDRIEARAMFHDARFDSQFPEPFYQGHKSTNVLNPVFKGQQLVPLNADQSA-- 566
Query: 117 PLFKAISNATAELKVALTTRFQYKTWGIKSK 147
L K + E+ V + R ++K K+K
Sbjct: 567 ELKKENATGVYEIDVKMYLRVRFKLGVFKTK 659
>TC211792 homologue to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein
B13-1-9, partial (48%)
Length = 421
Score = 36.2 bits (82), Expect = 0.009
Identities = 20/69 (28%), Positives = 36/69 (51%), Gaps = 6/69 (8%)
Frame = +3
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSLEIE----NPNK 64
++ I +GL+ L WL ++P+ + V + Q + N ++ Y++ + NPNK
Sbjct: 174 VVALIVIVGLVFLIFWLVVQPRYFKFHVTEADLTQFEYYPNNNTLHYNMVLNFTARNPNK 353
Query: 65 DSSIYYDDI 73
SIYYD +
Sbjct: 354 KLSIYYDKV 380
>BI699875 similar to PIR|E96668|E96 protein F1N19.3 [imported] - Arabidopsis
thaliana, partial (37%)
Length = 421
Score = 34.3 bits (77), Expect = 0.035
Identities = 18/74 (24%), Positives = 38/74 (51%), Gaps = 5/74 (6%)
Frame = +1
Query: 14 FIGFLGLLVLCLWLAL-RPKSPSYSVVFLSIEQHPGENGSIFYSLE----IENPNKDSSI 68
F+ FL ++V ++ + +P+ P +V + I NG++ ++ + NPN+ +
Sbjct: 28 FLIFLLIVVFIVYFTVFKPQDPKIAVSAVQIPSFSATNGTVNFTFSQYASVRNPNRGTFS 207
Query: 69 YYDDIILSFLYGQQ 82
+YD + YG+Q
Sbjct: 208 HYDSSLQLLYYGRQ 249
>TC222036 similar to PIR|E96668|E96668 protein F1N19.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(48%)
Length = 622
Score = 33.9 bits (76), Expect = 0.046
Identities = 18/74 (24%), Positives = 38/74 (51%), Gaps = 5/74 (6%)
Frame = +3
Query: 14 FIGFLGLLVLCLWLAL-RPKSPSYSVVFLSIEQHPGENGSIFYSLE----IENPNKDSSI 68
F+ FL ++V ++ + +P+ P +V + I NG++ ++ + NPN+ +
Sbjct: 84 FLIFLLIVVFIVYFTVFKPQYPKIAVSAIQIPSFSATNGTVNFTFSQYASVRNPNRGTFS 263
Query: 69 YYDDIILSFLYGQQ 82
+YD + YG+Q
Sbjct: 264 HYDSSLQLLYYGRQ 305
>TC218785 weakly similar to GB|BAB08954.1|9758412|AB006700 harpin-induced
protein-like {Arabidopsis thaliana;} , partial (28%)
Length = 993
Score = 33.1 bits (74), Expect = 0.079
Identities = 21/87 (24%), Positives = 44/87 (50%), Gaps = 5/87 (5%)
Frame = +2
Query: 20 LLVLCLWLALRPKSPSYSVVFLSIEQ-HPGENGSIFYSLEIE----NPNKDSSIYYDDII 74
+ ++ + + + P S + V S+ Q + N +++Y+L++ NPNK + +YY I
Sbjct: 194 MFIISIIVCISPSSVKFHVTDASLTQFNLTSNNTLYYNLKVNVTVRNPNKHTIVYYRRIT 373
Query: 75 LSFLYGQQEDKVGETTIGSFHQGTGKT 101
+ Y +++ G ++ F QG T
Sbjct: 374 VISWY--KDNAFGWVSLTPFDQGHKNT 448
>TC216389 weakly similar to UP|Q9M663 (Q9M663) Harpin inducing protein
(Hin1), partial (62%)
Length = 667
Score = 33.1 bits (74), Expect = 0.079
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 5/71 (7%)
Frame = +2
Query: 8 YVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ-HPGENGSIFYSLE----IENP 62
+ ++ I +G+ WL +RP + V ++ Q + N ++ Y L + NP
Sbjct: 173 FKLILTVIIIVGIAGFVFWLIVRPNVVKFHVTDATLTQFNYTANNTLHYDLALNITVRNP 352
Query: 63 NKDSSIYYDDI 73
NK IYYD I
Sbjct: 353 NKRLGIYYDRI 385
>TC208147 homologue to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein
B13-1-9, partial (53%)
Length = 567
Score = 32.7 bits (73), Expect = 0.10
Identities = 19/63 (30%), Positives = 34/63 (53%), Gaps = 6/63 (9%)
Frame = +3
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSLEI----ENPNK 64
L+ I +GL VL WL ++P+S + V + Q + N ++ Y++ + NPNK
Sbjct: 282 LVALIVLVGLAVLIFWLVVQPRSFKFHVTEADLTQFDYYTNNNTLHYNMVLNFTARNPNK 461
Query: 65 DSS 67
+S+
Sbjct: 462 NST 470
>TC215224
Length = 1219
Score = 32.3 bits (72), Expect = 0.13
Identities = 17/69 (24%), Positives = 33/69 (47%), Gaps = 5/69 (7%)
Frame = +2
Query: 20 LLVLCLWLALRPKSPSYSVVFLSIEQHPGENGSIFYS-----LEIENPNKDSSIYYDDII 74
L +L +W LRP P++++ +++ + S L NPN + +YYD +
Sbjct: 200 LTILLIWAILRPTKPTFTLQDVTVYAFNATIPNFLTSNFQVTLISRNPNDNIGVYYDRLE 379
Query: 75 LSFLYGQQE 83
+ +Y Q+
Sbjct: 380 IYVIYRSQQ 406
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,438,611
Number of Sequences: 63676
Number of extensions: 111103
Number of successful extensions: 661
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 656
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 658
length of query: 189
length of database: 12,639,632
effective HSP length: 92
effective length of query: 97
effective length of database: 6,781,440
effective search space: 657799680
effective search space used: 657799680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0358.5


