

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
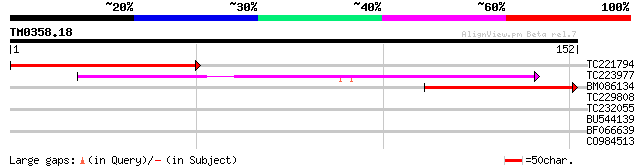
Query= TM0358.18
(152 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%) 70 3e-13
TC223977 weakly similar to UP|Q9FJS2 (Q9FJS2) Homeobox protein, ... 59 9e-10
BM086134 47 3e-06
TC229808 similar to UP|Q6K832 (Q6K832) Lipase class 3 protein-li... 28 2.2
TC232055 similar to UP|O49524 (O49524) Pherophorin - like protei... 27 3.8
BU544139 27 4.9
BF066639 26 6.4
CO984513 26 6.4
>TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%)
Length = 501
Score = 70.5 bits (171), Expect = 3e-13
Identities = 32/51 (62%), Positives = 39/51 (75%)
Frame = +3
Query: 1 ATCTTCGGLTSLGEMSYDEQLMKLENARLREAIERISGIVAKYAGKSTTSY 51
A+C CGG T++GEMS+DE ++LENARLRE I+RIS I AKY GK SY
Sbjct: 333 ASCPNCGGPTAIGEMSFDEHHLRLENARLREEIDRISAIAAKYVGKPVVSY 485
>TC223977 weakly similar to UP|Q9FJS2 (Q9FJS2) Homeobox protein, partial (7%)
Length = 447
Score = 58.9 bits (141), Expect = 9e-10
Identities = 38/141 (26%), Positives = 68/141 (47%), Gaps = 17/141 (12%)
Frame = +2
Query: 19 EQLMKLENARLREAIERISGIVAKYAGKSTTSYSSLLSQNYNQMPSSSRAFDLGVGNYGG 78
E +++ENARLRE +ER+ + +Y G+ + ++ + + DL + Y
Sbjct: 2 EHQLRIENARLREELERVCCLTTRYTGRPIQTMAA-------DPTLMAPSLDLDMNMYPR 160
Query: 79 DGNDLLRSS--------LPP---------ILADADKPIIVEVAVAAMEELVRLARVGHPL 121
+D + LPP +L + +K + +E+A ++M ELV++ + PL
Sbjct: 161 HFSDPIAPCTEMIPVPMLPPEASPFSEGGVLMEEEKSLALELAASSMAELVKMCQTNEPL 340
Query: 122 WVLSNNHNVETLNEEEYVREF 142
W+ S+ E LN EE+ R F
Sbjct: 341 WIQSSEGEREVLNFEEHARMF 403
>BM086134
Length = 428
Score = 47.4 bits (111), Expect = 3e-06
Identities = 19/41 (46%), Positives = 28/41 (67%)
Frame = +1
Query: 112 VRLARVGHPLWVLSNNHNVETLNEEEYVREFPRGTGSKPFG 152
+ +A++G PLW+ + + LNE+EY+R FPRG G KP G
Sbjct: 1 IGMAQMGEPLWLTTLDGTSTMLNEDEYIRSFPRGIGPKPSG 123
>TC229808 similar to UP|Q6K832 (Q6K832) Lipase class 3 protein-like, partial
(3%)
Length = 1019
Score = 27.7 bits (60), Expect = 2.2
Identities = 14/43 (32%), Positives = 24/43 (55%)
Frame = +3
Query: 40 VAKYAGKSTTSYSSLLSQNYNQMPSSSRAFDLGVGNYGGDGND 82
+A+ G S T++ SLL++ + M S S + + G DG+D
Sbjct: 174 IAEAYGSSETNFESLLTEEHLIMESMSDDDEYNSSSEGSDGDD 302
>TC232055 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(7%)
Length = 448
Score = 26.9 bits (58), Expect = 3.8
Identities = 18/60 (30%), Positives = 27/60 (45%)
Frame = -2
Query: 37 SGIVAKYAGKSTTSYSSLLSQNYNQMPSSSRAFDLGVGNYGGDGNDLLRSSLPPILADAD 96
SG + +AG + T+++ ++ AFD G GG G +LRS P D D
Sbjct: 363 SGTLRTFAGATLTAFAG---GGGGGGGGAAAAFDAGFLGGGGGGGGILRSVSPEETLDGD 193
>BU544139
Length = 424
Score = 26.6 bits (57), Expect = 4.9
Identities = 15/45 (33%), Positives = 23/45 (50%), Gaps = 1/45 (2%)
Frame = -3
Query: 104 AVAAMEELVRLARVGHPLW-VLSNNHNVETLNEEEYVREFPRGTG 147
++A + E +L PL+ ++ N HN+ LNE V F TG
Sbjct: 146 SLAIVAEAQKLLMTRAPLYYIVCNLHNLSLLNENGKVNSFKNHTG 12
>BF066639
Length = 353
Score = 26.2 bits (56), Expect = 6.4
Identities = 15/55 (27%), Positives = 32/55 (57%), Gaps = 1/55 (1%)
Frame = -2
Query: 38 GIVAKYAGKSTTSY-SSLLSQNYNQMPSSSRAFDLGVGNYGGDGNDLLRSSLPPI 91
G+V ++ +S++ S++LS + + SS GVG++ +G +L + + PP+
Sbjct: 268 GLVGQFLLRSSSIICSAILSSSSISVLRSSCLATCGVGSHSENGKELEQEASPPV 104
>CO984513
Length = 771
Score = 26.2 bits (56), Expect = 6.4
Identities = 15/39 (38%), Positives = 25/39 (63%), Gaps = 6/39 (15%)
Frame = -2
Query: 63 PSSSRAFDLGV---GNYGGDGNDLLRSSL---PPILADA 95
P+ S + DL + ++GG GN+LL SS+ PP+L+ +
Sbjct: 389 PAESSSEDLNMFQTNDHGGGGNNLLLSSVQQQPPLLSSS 273
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,374,013
Number of Sequences: 63676
Number of extensions: 70101
Number of successful extensions: 259
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 257
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 258
length of query: 152
length of database: 12,639,632
effective HSP length: 89
effective length of query: 63
effective length of database: 6,972,468
effective search space: 439265484
effective search space used: 439265484
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0358.18


