

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.7
(778 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
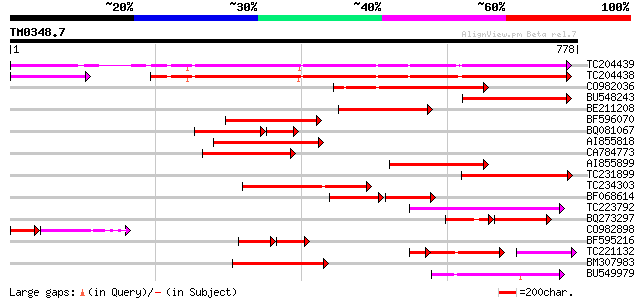
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 439 e-123
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 422 e-118
CO982036 221 1e-57
BU548243 185 8e-47
BE211208 166 3e-41
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 166 4e-41
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 116 3e-40
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 157 1e-38
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 156 3e-38
AI855899 similar to GP|2244960|emb| retrotransposon like protein... 155 8e-38
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 154 1e-37
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 153 3e-37
BF068614 similar to GP|27817858|db OJ1081_B12.7 {Oryza sativa (j... 92 3e-36
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 148 8e-36
BQ273297 95 2e-34
CO982898 94 4e-32
BF595216 82 2e-31
TC221132 weakly similar to UP|O23529 (O23529) RETROTRANSPOSON li... 82 4e-31
BM307983 130 3e-30
BU549979 120 2e-27
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 439 bits (1130), Expect = e-123
Identities = 269/785 (34%), Positives = 419/785 (53%), Gaps = 16/785 (2%)
Frame = +1
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERK+R + E +L +P + W A TA Y+ NR++ + Y G P
Sbjct: 2566 VERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPS 2745
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLDQSGRIYVSKDVLFH 121
K IFGS C+ K+ S +FL GY ++ R++ S+
Sbjct: 2746 VKHFHIFGSPCYILADREQRRKMDPKSDAGIFL-------GYSTNSRAYRVFNSRTRTVM 2904
Query: 122 EHRFPYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPSPAPISTELANPGPLSPQSE 181
E I+ + + P +
Sbjct: 2905 ES------------------------------------------INVVVDDLSPARKKDV 2958
Query: 182 ASDLQSQPSPIPTGSGLASTSQPAEHA-SSESAHQEMATSSGVHAASSASTVAVPVNAHP 240
D+++ +G +A ++ E+A +S+SA E S ++ S+ + HP
Sbjct: 2959 EEDVRT------SGDNVADAAKSGENAENSDSATDE----SNINQPDKRSSTRIQ-KMHP 3105
Query: 241 MQ-----------TRSKSGIIKPRLNPTLLLTHMEPTTVKQAMRDDKWLQAMKEEYNALM 289
+ TRS+ I ++ + ++ +EP VK+A+ D+ W+ AM+EE
Sbjct: 3106 KELIIGDPNRGVTTRSREVEI---VSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFK 3276
Query: 290 SNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPV 349
N W LVP P +G KWI++ K N +G I + KARLVA+GY+Q++G D+ ETF+PV
Sbjct: 3277 RNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPV 3456
Query: 350 VKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFE---HKDKTLVCKL 406
+ ++RL+L +A + L Q+DV +AFLNG L EEVY+ QP GF H D V +L
Sbjct: 3457 ARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDH--VYRL 3630
Query: 407 HKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGD 466
KALYGLKQAPRAW+ RL E L Q G++ D +LF + IYVDDI+ G
Sbjct: 3631 KKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGM 3810
Query: 467 SMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADA 526
S +++ ++ S F + +G+L YFLG+QV + + S+ L+Q++Y +++ K M +A
Sbjct: 3811 SNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMED-SIFLSQSRYAKNIVKKFGMENA 3987
Query: 527 APISTPMQFGAKLTK-HGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPH 585
+ TP KL+K GTS+ D + YRS++G+L Y T +RP+I+YAV ++ ++P
Sbjct: 3988 SHKRTPAPTHLKLSKDEAGTSV-DQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPK 4164
Query: 586 EEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFL 645
H VKRIL+Y+ GT +G++ CS P+ L+ +CDADW DDR+STSG C +L
Sbjct: 4165 ISHLTQVKRILKYVNGTSDYGIMYCHCS--NPM-LVGYCDADWAGSADDRKSTSGGCFYL 4335
Query: 646 GPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVLCDNMST 705
G NLISW +KKQ V+ S+ EAEY + ++ ++L+W++ +L E + V T+ CDNMS
Sbjct: 4336 GNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSA 4515
Query: 706 VLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMR 765
+ ++ NP+ H+RTKH+++ ++R+ V K + ++HV +E Q ADIFTKAL +F +R
Sbjct: 4516 INISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFEKLR 4695
Query: 766 DKLNV 770
KL +
Sbjct: 4696 GKLGI 4710
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 422 bits (1084), Expect = e-118
Identities = 232/593 (39%), Positives = 358/593 (60%), Gaps = 16/593 (2%)
Frame = +1
Query: 194 TGSGLASTSQPAEHA-SSESAHQEMATSSGVHAASSASTVAVPVNAHPMQ---------- 242
+G +A T++ AE+A +S+SA E ++ ++ + HP +
Sbjct: 2980 SGDNVADTAKSAENAENSDSATDE----PNINQPDKRPSIRIQ-KMHPKELIIGDPNRGV 3144
Query: 243 -TRSKSGIIKPRLNPTLLLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPS 301
TRS+ I ++ + ++ +EP VK+A+ D+ W+ AM+EE N W LVP P
Sbjct: 3145 TTRSREIEI---VSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPE 3315
Query: 302 NRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSL 361
+G KWI++ K N +G I + KARLVA+GY+Q++G D+ ETF+PV + ++RL+L +
Sbjct: 3316 GTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGV 3495
Query: 362 AISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGF---EHKDKTLVCKLHKALYGLKQAPR 418
A + L Q+DV +AFLNG L EE Y+ QP GF H D V +L KALYGLKQAPR
Sbjct: 3496 ACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDH--VYRLKKALYGLKQAPR 3669
Query: 419 AWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKL 478
AW+ RL E L Q G++ D +LF + IYVDDI+ G S +++ ++
Sbjct: 3670 AWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQM 3849
Query: 479 HSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQFGAK 538
S F + +G+L YFLG+QV + + S+ L+Q+KY +++ K M +A+ TP K
Sbjct: 3850 QSEFEMSLVGELTYFLGLQVKQMED-SIFLSQSKYAKNIVKKFGMENASHKRTPAPTHLK 4026
Query: 539 LTK-HGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRILR 597
L+K GTS+ D + YRS++G+L Y T +RP+I+YAV ++ ++P H VKRIL+
Sbjct: 4027 LSKDEAGTSV-DQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLNQVKRILK 4203
Query: 598 YLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQ 657
Y+ GT +G++ CS + L+ +CDADW DDR+STSG C +LG NLISW +KKQ
Sbjct: 4204 YVNGTSDYGIMYCHCSDSM---LVGYCDADWAGSADDRKSTSGGCFYLGTNLISWFSKKQ 4374
Query: 658 TLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVLCDNMSTVLLTHNPILHTR 717
V+ S+ EAEY + ++ ++L+W++ +L E + V T+ CDNMS + ++ NP+ H+R
Sbjct: 4375 NCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQHSR 4554
Query: 718 TKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
TKH+++ ++R+ V K + ++HV +E Q ADIFTKAL +F +R KL +
Sbjct: 4555 TKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKALDANQFEKLRGKLGI 4713
Score = 57.8 bits (138), Expect = 2e-08
Identities = 33/110 (30%), Positives = 50/110 (45%)
Frame = +1
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERK+R + E +L +P + W A TA Y+ NR++ + Y G P
Sbjct: 2569 VERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPT 2748
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLDQSGR 111
K IFGS C+ K+ S +FLGYS++ + Y+ + R
Sbjct: 2749 VKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTR 2898
>CO982036
Length = 674
Score = 221 bits (563), Expect = 1e-57
Identities = 114/213 (53%), Positives = 149/213 (69%)
Frame = -2
Query: 445 YNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANG 504
Y + + +Y+L+YVD II+TG S LIQ LT+KL+S F LK LG+LDYF+ ++V + +
Sbjct: 673 YKTHILTVYLLVYVD-IIITGSSCTLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPD- 500
Query: 505 SLLLNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYAT 564
LL + I ++ + A PIS+PM KL+K PT YRSVVGALQY T
Sbjct: 499 -LLFSLRTSIFEIFCRKPR*QAQPISSPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTT 323
Query: 565 ITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFC 624
+ RPEIS+AVNKVCQF+S+P + HW VKRILRYLKG++++G+ L+P +QPLP+ FC
Sbjct: 322 VIRPEISFAVNKVCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFC 143
Query: 625 DADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQ 657
DADW S DD+RSTSG+ VFLGPNLISW KQ
Sbjct: 142 DADWASAVDDKRSTSGAAVFLGPNLISWWXXKQ 44
>BU548243
Length = 599
Score = 185 bits (469), Expect = 8e-47
Identities = 87/149 (58%), Positives = 110/149 (73%)
Frame = -1
Query: 622 AFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLW 681
A CDA W SD DD RST GS +FLGPNLISW ++KQ + A+SSTEAEYRS+A T+AEL W
Sbjct: 596 ALCDAGWASDVDDHRSTLGSAIFLGPNLISWWSRKQQVTAQSSTEAEYRSIAQTSAELTW 417
Query: 682 VESLLTELKIAFTVPTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQH 741
+++LL EL+I FT P +LCDN S V + HN + H+RTKHME+D+FFV EKV +K L + H
Sbjct: 416 IQALLMELQIPFTPPVILCDNKSAVAIAHNLVFHSRTKHMEIDVFFVHEKVLSKQLQIFH 237
Query: 742 VPSEHQRADIFTKALSPTRFLLMRDKLNV 770
+P+ Q A I TK LS RF ++ KL V
Sbjct: 236 IPALDQWAGILTKPLSSARFTFLKSKLTV 150
>BE211208
Length = 413
Score = 166 bits (421), Expect = 3e-41
Identities = 84/129 (65%), Positives = 101/129 (78%)
Frame = +2
Query: 452 IYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQT 511
+Y+L+YVDDII+TG S LIQ L L+S F+LKQLGQLDYFLG++V H GS+LL Q+
Sbjct: 26 VYLLVYVDDIIITGRSNYLIQSLVHHLNSNFSLKQLGQLDYFLGIEVHHTPTGSVLLTQS 205
Query: 512 KYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEIS 571
KYI DLL K +MA+A PIS+PM +L+K+G L DPT YRSVVGALQY TITRPEIS
Sbjct: 206 KYICDLLHKTDMAEAKPISSPMVTNLRLSKNGDDLLSDPTMYRSVVGALQYPTITRPEIS 385
Query: 572 YAVNKVCQF 580
+A NKVCQF
Sbjct: 386 FAANKVCQF 412
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 166 bits (420), Expect = 4e-41
Identities = 77/133 (57%), Positives = 98/133 (72%), Gaps = 1/133 (0%)
Frame = -2
Query: 297 VPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVR 356
VPLP + VGC+W+Y VK P G +++ KARLVAKGY+QV G DY +TFSPV K TVR
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 357 LILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGF-EHKDKTLVCKLHKALYGLKQ 415
L L++A WPL Q+D+ NAFL+G LEE++YM QPPGF + LVCKLH++LYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 416 APRAWFHRLKEVL 428
+PRAWF + V+
Sbjct: 46 SPRAWFGKFSHVV 8
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 116 bits (290), Expect(2) = 3e-40
Identities = 53/98 (54%), Positives = 74/98 (75%)
Frame = +1
Query: 254 LNPTLLLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYR 313
L+PTLL T EP+TVKQA+ W QAM+ +++ALM N T +L LPS + A+ CKW++R
Sbjct: 1 LHPTLLFTTAEPSTVKQALISPPWRQAMQADFDALMENKTLTLTSLPSGKAAIDCKWVFR 180
Query: 314 VKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVK 351
+KEN G++N+Y++RLVAKG+ G DYSETFSPV++
Sbjct: 181 IKENLYGTLNRYRSRLVAKGFHLKFGCDYSETFSPVIE 294
Score = 68.6 bits (166), Expect(2) = 3e-40
Identities = 29/44 (65%), Positives = 38/44 (85%)
Frame = +3
Query: 353 ITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFE 396
+T+RLIL +A++ WPLQQ+D+NNAFL+G+L EEVYM Q PGFE
Sbjct: 297 VTIRLILFIALTNHWPLQQVDINNAFLHGLLTEEVYMVQLPGFE 428
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 157 bits (398), Expect = 1e-38
Identities = 78/152 (51%), Positives = 103/152 (67%), Gaps = 1/152 (0%)
Frame = -3
Query: 280 AMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQG 339
AM+EE N N W LV P N +G KW++R K + G I + KARLVAKGY+Q +G
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 340 FDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKD 399
DY ET++PV + +R++L+ + L Q+DV +AFLNG+++EEVY+ QPPGFE D
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 400 K-TLVCKLHKALYGLKQAPRAWFHRLKEVLLQ 430
K T V KL KALYGLKQAPRAW+ R+ LL+
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 156 bits (395), Expect = 3e-38
Identities = 70/128 (54%), Positives = 94/128 (72%)
Frame = +3
Query: 265 PTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINK 324
P+T+++A+ W QAM +E AL +NGTW LVPLP + VGC+W+Y VK P+G +++
Sbjct: 21 PSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNGKVDR 200
Query: 325 YKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLE 384
KARLVAKGY+QV G +Y +TFSPV TVRL L++A R WPL Q+D+ NAFL+G LE
Sbjct: 201 LKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLHGDLE 380
Query: 385 EEVYMTQP 392
E++YM QP
Sbjct: 381 EDIYMEQP 404
>AI855899 similar to GP|2244960|emb| retrotransposon like protein
{Arabidopsis thaliana}, partial (18%)
Length = 418
Score = 155 bits (391), Expect = 8e-38
Identities = 76/136 (55%), Positives = 95/136 (68%)
Frame = +1
Query: 522 NMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFL 581
NM D ISTPM KL+K G L + +YR +VGALQY T+TRP I+Y VNKV +F+
Sbjct: 10 NMLDCNGISTPMVSSYKLSKFGSELLPNAHQYRDIVGALQYVTLTRPNIAYNVNKVSEFM 189
Query: 582 SDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGS 641
S P + + VKRILRYL GT+T G+LLQP M + L A+ D DWGSDP + RSTSGS
Sbjct: 190 SSPLQSY*LTVKRILRYLSGTVTQGLLLQPAHMDAKISLRAYNDLDWGSDPAEMRSTSGS 369
Query: 642 CVFLGPNLISWTAKKQ 657
C+F G NLI+W++KKQ
Sbjct: 370 CIFSGSNLIAWSSKKQ 417
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (30%)
Length = 687
Score = 154 bits (390), Expect = 1e-37
Identities = 74/154 (48%), Positives = 101/154 (65%), Gaps = 1/154 (0%)
Frame = +2
Query: 620 LLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAEL 679
L +CDADW P DRRSTSG CVF+G NL+SW +KKQT+VARSS EAEYRS+A T EL
Sbjct: 17 LSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCEL 196
Query: 680 LWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLV 738
+W++ L EL+ + L CDN + + + NP+ H RTKH+E+D F+REK+ +K +V
Sbjct: 197 MWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSKEIV 376
Query: 739 VQHVPSEHQRADIFTKALSPTRFLLMRDKLNVVD 772
+ + S Q DI TK+L + ++ KL D
Sbjct: 377 TEFIGSNDQPVDILTKSLRGPKIQIVCSKLGAYD 478
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 153 bits (386), Expect = 3e-37
Identities = 85/179 (47%), Positives = 115/179 (63%), Gaps = 2/179 (1%)
Frame = +1
Query: 320 GSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFL 379
G+I+++KARLVAK Y+QV G DY+ TFSPV K V L+ S+A+ WPL +D NAFL
Sbjct: 28 GTIDQFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFL 207
Query: 380 NGVLEEEVYMTQPPGF--EHKDKTLVCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASK 437
+G LEEEVYM QP GF + + +VC+L ++ YGLKQ+PRAW + + + +
Sbjct: 208 HGYLEEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAWPFLYCGAAI--WYDSHE 381
Query: 438 CDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGV 496
D S+F +SP GCIY+++YVDDI +TG I L L F K LG+L YFLG+
Sbjct: 382 ADHSVFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>BF068614 similar to GP|27817858|db OJ1081_B12.7 {Oryza sativa (japonica
cultivar-group)}, partial (3%)
Length = 413
Score = 92.4 bits (228), Expect(2) = 3e-36
Identities = 45/69 (65%), Positives = 51/69 (73%)
Frame = +1
Query: 516 DLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVN 575
DLL K MA+A PIS+PM KL+K G DPT Y SVVGALQYAT+TRPE SY+VN
Sbjct: 202 DLLPKTKMAEAQPISSPMVSSCKLSKSGSDLFQDPTLYGSVVGALQYATLTRPEFSYSVN 381
Query: 576 KVCQFLSDP 584
KVCQF+S P
Sbjct: 382 KVCQFMSQP 408
Score = 79.0 bits (193), Expect(2) = 3e-36
Identities = 37/75 (49%), Positives = 55/75 (73%)
Frame = +2
Query: 439 DPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQV 498
DPSLF Y +++L+YVDDI +TG+ ++LIQQL ++L+S FALKQLG LDYFLG++V
Sbjct: 2 DPSLFVYKQQQYTVFLLVYVDDITITGNCVLLIQQLISQLNSQFALKQLGLLDYFLGIEV 181
Query: 499 THLANGSLLLNQTKY 513
+L + + + K+
Sbjct: 182 KYLPDKGISCPRLKW 226
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (7%)
Length = 804
Score = 148 bits (374), Expect = 8e-36
Identities = 76/214 (35%), Positives = 128/214 (59%), Gaps = 1/214 (0%)
Frame = +1
Query: 549 DPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVL 608
D TE+R ++G+L+Y +RP I +AV+ + +F+ P H +A KR+LR +KGTI GVL
Sbjct: 10 DVTEFRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVL 189
Query: 609 LQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAE 668
+ + LL + D+DW DP+ +ST G ++ ++KKQ ++A S+ EAE
Sbjct: 190 FPFKAKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAE 369
Query: 669 YRSLANTTAELLWVESLLTELKIAFTVP-TVLCDNMSTVLLTHNPILHTRTKHMEMDLFF 727
Y + + + +W+ +LL ELK+ P +L DN S + L +P LH R+KH+E+ +
Sbjct: 370 YVAASLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHY 549
Query: 728 VREKVQAKSLVVQHVPSEHQRADIFTKALSPTRF 761
+R++V ++ V++ +E Q AD+ TK + +RF
Sbjct: 550 IRDQVSKGNVTVEYCKAEEQLADLMTKPIQVSRF 651
>BQ273297
Length = 420
Score = 94.7 bits (234), Expect(2) = 2e-34
Identities = 46/78 (58%), Positives = 59/78 (74%)
Frame = +2
Query: 666 EAEYRSLANTTAELLWVESLLTELKIAFTVPTVLCDNMSTVLLTHNPILHTRTKHMEMDL 725
E E+RS+ A++ ++SLL+EL++A + P +LCDN STV L HN ILH+RTKHME+DL
Sbjct: 185 ETEHRSMTLIVAKVT*IQSLLSELQVAHSTPLILCDNTSTVSLAHNLILHSRTKHMELDL 364
Query: 726 FFVREKVQAKSLVVQHVP 743
FFVREKV K L V HVP
Sbjct: 365 FFVREKVLNKKLDVVHVP 418
Score = 70.5 bits (171), Expect(2) = 2e-34
Identities = 37/66 (56%), Positives = 46/66 (69%)
Frame = +1
Query: 599 LKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQT 658
LKGTI+ G+ Q S+ P+PL A+C +DW SDPD RRS FLGPNLI +KKQ+
Sbjct: 1 LKGTISWGLHFQATSLQHPIPLHAYCGSDWASDPDYRRS------FLGPNLIC*WSKKQS 162
Query: 659 LVARSS 664
+VARSS
Sbjct: 163 VVARSS 180
>CO982898
Length = 816
Score = 93.6 bits (231), Expect(2) = 4e-32
Identities = 52/124 (41%), Positives = 72/124 (57%)
Frame = -1
Query: 43 TTTLQGASPYFKLYGNHPDFKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKG 102
T ++ A PY +L+ D+ ++FG ACFP +RPYN K + S EC+FLG +SHK
Sbjct: 594 TVSINFAVPYTQLFNQMLDYSFPRVFGCACFPLIRPYNIQKFNFKSHECIFLGNYTSHKN 415
Query: 103 YKCLDQSGRIYVSKDVLFHEHRFPYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPS 162
YKCL +G++ VSKDV+F+E FPY LF P SS + +P+ PQP
Sbjct: 414 YKCLAPAGKL-VSKDVIFNEI*FPYKELF------LPDSS------TCIPV-----PQPI 289
Query: 163 PAPI 166
PA +
Sbjct: 288 PATV 277
Score = 63.9 bits (154), Expect(2) = 4e-32
Identities = 28/40 (70%), Positives = 34/40 (85%)
Frame = -3
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRM 41
VERK RHIV+ GL+LLSHAS+PL FW HAF+T +LINR+
Sbjct: 718 VERKQRHIVKFGLSLLSHASLPLKFWGHAFITPVFLINRL 599
>BF595216
Length = 421
Score = 82.4 bits (202), Expect(2) = 2e-31
Identities = 34/45 (75%), Positives = 41/45 (90%)
Frame = +3
Query: 367 WPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDKTLVCKLHKALY 411
WP+QQ+DVNNAFLNG+L+EEVYM QPPGF+ K+LVCKLHKA+Y
Sbjct: 285 WPIQQLDVNNAFLNGILQEEVYMQQPPGFDSTTKSLVCKLHKAIY 419
Score = 72.4 bits (176), Expect(2) = 2e-31
Identities = 33/50 (66%), Positives = 43/50 (86%)
Frame = +1
Query: 315 KENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAIS 364
KEN DG++NKYKA+LVAKG+ Q G DY ETFS V+KPIT+RL+L+LA++
Sbjct: 124 KENSDGTVNKYKAKLVAKGFHQQYGTDYIETFSLVIKPITMRLLLTLAVT 273
>TC221132 weakly similar to UP|O23529 (O23529) RETROTRANSPOSON like protein,
partial (5%)
Length = 799
Score = 81.6 bits (200), Expect(3) = 4e-31
Identities = 42/107 (39%), Positives = 65/107 (60%)
Frame = +1
Query: 572 YAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSD 631
++++ QF+ DP + +A KR+LRYLKGTI G+ L+ + L AF DA+W +
Sbjct: 115 HSLSTSSQFMKDPTKIRMQATKRVLRYLKGTIDFGLQLRS---SPDQHLRAFYDANWVDN 285
Query: 632 PDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAE 678
D RST V+ G ++ISW+ KKQ+++ +SST+ EY + T E
Sbjct: 286 TSDIRSTGAYVVYFGLSVISWSCKKQSIIDKSSTKVEYHKITTTIIE 426
Score = 64.7 bits (156), Expect(3) = 4e-31
Identities = 34/82 (41%), Positives = 46/82 (55%)
Frame = +2
Query: 696 PTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKA 755
PT+ N+ + L NP+ H KH+ +D FV++ V K L V HVPS H D+FTKA
Sbjct: 449 PTMYSYNIGAMYLCANPVFHLCMKHLTIDHLFVQDLVANKQLRVSHVPSCH*HVDLFTKA 628
Query: 756 LSPTRFLLMRDKLNVVDKHLML 777
L +R M DK+ VV +L
Sbjct: 629 LVSSRHKFMMDKIGVVSTTTIL 694
Score = 28.1 bits (61), Expect(3) = 4e-31
Identities = 11/29 (37%), Positives = 20/29 (68%)
Frame = +3
Query: 549 DPTEYRSVVGALQYATITRPEISYAVNKV 577
D Y +V +LQY ++T P+I++ +NK+
Sbjct: 48 DGIVYCQLVDSLQYLSLTCPDIAFPINKL 134
>BM307983
Length = 406
Score = 130 bits (326), Expect = 3e-30
Identities = 69/134 (51%), Positives = 86/134 (63%), Gaps = 2/134 (1%)
Frame = +2
Query: 306 VGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISR 365
VGC+WIY VK D ++++YKARLVAKGY Q G DY ETF+ K I ++
Sbjct: 2 VGCRWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQ 181
Query: 366 -GWPLQQIDVNNAFLNGVLEEEVYMTQPPGF-EHKDKTLVCKLHKALYGLKQAPRAWFHR 423
GW + Q DV NAFL+G LEEEVYM PPG+ VC+L KALYGLKQ+PRAWF R
Sbjct: 182 FGWEMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGR 361
Query: 424 LKEVLLQFGFKASK 437
+ +L G+K S+
Sbjct: 362 FTQAMLSLGYKQSQ 403
>BU549979
Length = 615
Score = 120 bits (301), Expect = 2e-27
Identities = 59/186 (31%), Positives = 107/186 (56%), Gaps = 3/186 (1%)
Frame = -1
Query: 579 QFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRST 638
++ S+P +HWK K+++RYL+GT + ++ + T L ++ + D+D+ D RRST
Sbjct: 606 RYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYK---QTNCLEVIGYSDSDFAGCVDSRRST 436
Query: 639 SGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTV 698
SG L ++SW + KQTL+A S+ E E+ T+ +W++S ++ L++ ++
Sbjct: 435 SGYIFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRP 256
Query: 699 L---CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKA 755
L CDN + V + N R+KH+++ +RE+V+ K +V++HV +E D TK
Sbjct: 255 LKLYCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTELMIVDPLTKG 76
Query: 756 LSPTRF 761
++P F
Sbjct: 75 MTPKNF 58
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,969,307
Number of Sequences: 63676
Number of extensions: 612780
Number of successful extensions: 5879
Number of sequences better than 10.0: 542
Number of HSP's better than 10.0 without gapping: 4821
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5409
length of query: 778
length of database: 12,639,632
effective HSP length: 105
effective length of query: 673
effective length of database: 5,953,652
effective search space: 4006807796
effective search space used: 4006807796
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0348.7


