

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
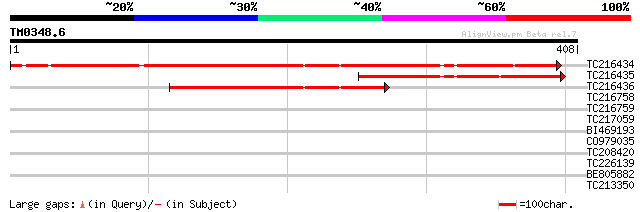
Query= TM0348.6
(408 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216434 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Frag... 397 e-111
TC216435 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Frag... 160 7e-40
TC216436 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Frag... 149 2e-36
TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase... 32 0.67
TC216759 similar to PIR|T05768|T05768 subtilisin-like proteinase... 30 2.6
TC217059 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, parti... 29 4.4
BI469193 similar to PIR|T05768|T05 subtilisin-like proteinase (E... 29 4.4
CO979035 28 7.4
TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%) 28 9.7
TC226139 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, ... 28 9.7
BE805882 28 9.7
TC213350 similar to UP|Q880Q7 (Q880Q7) Conserved domain protein,... 28 9.7
>TC216434 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (96%)
Length = 1436
Score = 397 bits (1020), Expect = e-111
Identities = 217/398 (54%), Positives = 292/398 (72%), Gaps = 1/398 (0%)
Frame = +1
Query: 1 MEIQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDEL 60
M++++S S Q+DVALS +K L SKE ++KNL++SPLSL+ LS++AAGS+G TLD+L
Sbjct: 10 MDLRESISN--QTDVALSISKLLLSKEA-RDKNLVYSPLSLHVVLSIIAAGSKGPTLDQL 180
Query: 61 LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLL 120
LSFLR S DHLN++ SQ + V SD PA P LSF +G++++ ++SL F++L+
Sbjct: 181 LSFLRSKSTDHLNSFASQLFAVVLSDASPAGGP---RLSFADGVWVEQSLSLLPSFKQLV 351
Query: 121 STHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQ 180
S Y A+LAS+DF + V ++VNS E+E+NGL+ LLPPG+V + T+LIFANAL F+
Sbjct: 352 SADYKATLASVDFQTKAVEVANEVNSWAEKETNGLVKDLLPPGSVDSSTRLIFANALYFK 531
Query: 181 GMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D +T F+LL G S++VP+MT+ +K Q++RAFDGFK+L LPYKQG D+ R
Sbjct: 532 GAWNEKFDSSITKDYDFHLLDGRSIRVPFMTS-RKNQFIRAFDGFKVLGLPYKQGEDK-R 705
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+F+M FLP+ +DGL AL +KL+SE GFL+ KLP+ K+ V FR+P+F ISF FEASNVL
Sbjct: 706 QFTMYFFLPETKDGLLALAEKLASESGFLERKLPNNKLEVGDFRIPRFKISFGFEASNVL 885
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQ 359
KE+GVV PFS T+MV+ ++ NLFV IFHK+FIEVNE+GT+A AAT + +
Sbjct: 886 KELGVVLPFSV--GGLTEMVD-SAVGQNLFVSDIFHKSFIEVNEEGTEAAAATAATIQFG 1056
Query: 360 CARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
CA IDF+ADHPFLFLIRED TGT+LF+GQVLNP
Sbjct: 1057CAM-FPTEIDFVADHPFLFLIREDLTGTVLFIGQVLNP 1167
>TC216435 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (37%)
Length = 820
Score = 160 bits (406), Expect = 7e-40
Identities = 90/149 (60%), Positives = 110/149 (73%)
Frame = +2
Query: 252 DGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPM 311
DGL AL +KL+SE GFL+ KLP++KV V FR+P+F ISF FE SNVLKE+GVV PFS
Sbjct: 11 DGLLALAEKLASESGFLERKLPNQKVEVGDFRIPRFKISFGFEVSNVLKELGVVLPFSV- 187
Query: 312 DAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFI 371
T+MV+ + NL V +IFHK+FIEVNE+GT+A AAT SA R + IDF+
Sbjct: 188 -GGLTEMVD-SPVGQNLCVSNIFHKSFIEVNEEGTEAAAAT-SATIRLRSAMLPTKIDFV 358
Query: 372 ADHPFLFLIREDFTGTILFVGQVLNPLDG 400
ADHPFLFLIRED TGT+LF+GQVL+P G
Sbjct: 359 ADHPFLFLIREDLTGTVLFIGQVLDPRAG 445
>TC216436 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (40%)
Length = 489
Score = 149 bits (376), Expect = 2e-36
Identities = 79/159 (49%), Positives = 112/159 (69%), Gaps = 1/159 (0%)
Frame = +2
Query: 116 FRRLLSTHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFAN 175
F++L+ Y A+LAS+DF + V ++VNS E+E+NGL+ LLPPG+V N TKLIFAN
Sbjct: 17 FKQLVRAQYKATLASVDFQTKAVEVTNEVNSWAEKETNGLVKDLLPPGSVDNSTKLIFAN 196
Query: 176 ALRFQGMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQG 234
AL +G W D +T +LL+G+SVKVP+MT+ KK Q++ A + FK+L L YKQG
Sbjct: 197 ALWCKGAWNEKFDASITKHYDLHLLNGSSVKVPFMTS-KKKQFIMA*NSFKVLGLSYKQG 373
Query: 235 RDRQRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLP 273
D+ R+F+M FLP+ +DGL AL +KL+ + GFL+ +LP
Sbjct: 374 EDK-RQFTMYFFLPETKDGLLALAEKLAFDSGFLERRLP 487
>TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(35%)
Length = 796
Score = 31.6 bits (70), Expect = 0.67
Identities = 27/96 (28%), Positives = 47/96 (48%), Gaps = 2/96 (2%)
Frame = +3
Query: 117 RRLLSTHYNASLASLDFN--LRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFA 174
++ +S+ N + A+LDF + G + S + NGL Q+L P + ++ A
Sbjct: 459 KKYISSSTNPT-ATLDFKGTILGIKPAPVIASFSARGPNGLNPQILKPDFIAPGVNILAA 635
Query: 175 NALRFQGMWKHTLDGLTHVSSFNLLSGTSVKVPYMT 210
Q + LD T + FN+LSGTS+ P+++
Sbjct: 636 WT---QAVGPTGLDSDTRRTEFNILSGTSMACPHVS 734
>TC216759 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(24%)
Length = 542
Score = 29.6 bits (65), Expect = 2.6
Identities = 25/96 (26%), Positives = 47/96 (48%), Gaps = 2/96 (2%)
Frame = +3
Query: 117 RRLLSTHYNASLASLDFN--LRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFA 174
++ +S+ N + A+LDF + G + S + NGL ++L P + ++ A
Sbjct: 252 KKYISSSKNPT-ATLDFKGTILGIKPAPVIASFSARGPNGLNPEILKPDLIAPGVNILAA 428
Query: 175 NALRFQGMWKHTLDGLTHVSSFNLLSGTSVKVPYMT 210
+ + LD T + FN+LSGTS+ P+++
Sbjct: 429 WT---EAVGPTGLDSDTRRTEFNILSGTSMACPHVS 527
>TC217059 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, partial (9%)
Length = 1565
Score = 28.9 bits (63), Expect = 4.4
Identities = 19/52 (36%), Positives = 24/52 (45%), Gaps = 4/52 (7%)
Frame = -2
Query: 77 SQGISPVFSDEDPASSP--SQHHLSFTNGM--FIDTTVSLSYPFRRLLSTHY 124
S SP+FS P S P HH + M F T + SYPF + + HY
Sbjct: 1078 SAASSPLFSSS*PLSCPLTQNHHYNPHQKMYTFSQTFYAFSYPFPPVYAYHY 923
>BI469193 similar to PIR|T05768|T05 subtilisin-like proteinase (EC 3.4.21.-)
- Arabidopsis thaliana, partial (17%)
Length = 401
Score = 28.9 bits (63), Expect = 4.4
Identities = 20/67 (29%), Positives = 32/67 (46%)
Frame = +3
Query: 144 VNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDGLTHVSSFNLLSGTS 203
V S + NGL ++L P ++ A G+ LD T + FN+LSGTS
Sbjct: 54 VASFSARGPNGLSLEILKPDLTAPGVNILAAWT---GGVGPSGLDSDTRRTEFNILSGTS 224
Query: 204 VKVPYMT 210
+ P+++
Sbjct: 225 MACPHVS 245
>CO979035
Length = 347
Score = 28.1 bits (61), Expect = 7.4
Identities = 15/58 (25%), Positives = 26/58 (43%)
Frame = -1
Query: 313 AHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDF 370
A F M+N S + ++ S+FH ++ + + W L QC +H +I F
Sbjct: 347 AAFLSMLNPLSXAYKFYIISLFHATLLQKQSLSSYLSLLPW*NLNTQCIALYHLNIYF 174
>TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%)
Length = 1523
Score = 27.7 bits (60), Expect = 9.7
Identities = 14/51 (27%), Positives = 24/51 (46%), Gaps = 3/51 (5%)
Frame = -2
Query: 329 FVQSIFHKAFIEVNEKGTKATAA---TWSALARQCARDHHPSIDFIADHPF 376
++ S+ AF+++ K +AA WSA + C D P ++ PF
Sbjct: 427 YLGSLLKLAFLQIQRVEAKTSAAFHPQWSAQSHLCESDSAPRLETAHKQPF 275
>TC226139 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(79%)
Length = 519
Score = 27.7 bits (60), Expect = 9.7
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = +3
Query: 137 GDSVLHDVNSLIEQESNGLIT 157
GD+ LHD +L +Q+ NGLI+
Sbjct: 366 GDTELHDAFNLYDQDKNGLIS 428
>BE805882
Length = 413
Score = 27.7 bits (60), Expect = 9.7
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 4/65 (6%)
Frame = +2
Query: 89 PASSPS----QHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASLDFNLRGDSVLHDV 144
P+ SPS HLSF + T+SLS R +L+ + S +SL L VLH++
Sbjct: 80 PSPSPSPPTISDHLSFLPAAILTLTLSLSSEDREVLAYLISCSPSSLQLQLL--PVLHEL 253
Query: 145 NSLIE 149
+E
Sbjct: 254 LGPLE 268
>TC213350 similar to UP|Q880Q7 (Q880Q7) Conserved domain protein, partial
(6%)
Length = 863
Score = 27.7 bits (60), Expect = 9.7
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = +2
Query: 66 FDSIDHLNTYFSQGISPVF 84
FD +H+NTY SQ SP+F
Sbjct: 38 FDPFNHINTYASQPHSPLF 94
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,870,211
Number of Sequences: 63676
Number of extensions: 221429
Number of successful extensions: 1101
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1086
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1089
length of query: 408
length of database: 12,639,632
effective HSP length: 100
effective length of query: 308
effective length of database: 6,272,032
effective search space: 1931785856
effective search space used: 1931785856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0348.6


