

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
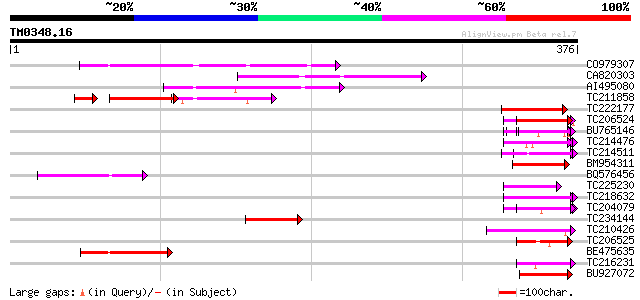
Query= TM0348.16
(376 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO979307 74 1e-13
CA820303 72 3e-13
AI495080 72 5e-13
TC211858 41 6e-13
TC222177 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich prot... 50 1e-06
TC206524 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 49 3e-06
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 49 4e-06
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 47 2e-05
TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protei... 46 2e-05
BM954311 weakly similar to GP|21322711|e pherophorin-dz1 protein... 46 3e-05
BQ576456 45 5e-05
TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein ... 45 7e-05
TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wal... 44 1e-04
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 42 3e-04
TC234144 42 3e-04
TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protei... 42 3e-04
TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (... 42 5e-04
BE475635 42 5e-04
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 42 6e-04
BU927072 42 6e-04
>CO979307
Length = 553
Score = 73.6 bits (179), Expect = 1e-13
Identities = 61/175 (34%), Positives = 87/175 (48%), Gaps = 2/175 (1%)
Frame = -3
Query: 47 HLVGATLFADKSGGHSFSARWIDMFQNLERVSEFAWGAMALATLYDQLGQSLRSGVKQMG 106
HL+ + +FADKS H A ++ NL + G AL LYD L KQ G
Sbjct: 551 HLISSMIFADKSLAHVHVA-YL*YLSNLTACHK*V*GVAALVYLYDHLSYVSLDDNKQCG 375
Query: 107 GYTSLLLAWVFEHFPDRLVRRYVSP-DYIEDQARACRWKE-SRSGHARLDERRVLLDDLT 164
Y +LL+A VFEH P Y +P DY + RW + HA L + R ++++
Sbjct: 374 DYMTLLMA*VFEHLPS---VGYSNPEDYSDGDPLNTRWNPLGGTRHALLVKER--MNNIH 210
Query: 165 EDDVIWTPYEAHREWRQRDERALFSGYIQCPYPPAVRPHLPERVMRQFGYIQTIP 219
+ I PYE ++ R E+ L GY +C +R +L +RV RQF ++QTIP
Sbjct: 209 MNVFI*CPYE*NKLVRPFVEQTL*FGYFKCRID--MRSYLLKRVRRQFCHVQTIP 51
>CA820303
Length = 421
Score = 72.4 bits (176), Expect = 3e-13
Identities = 44/125 (35%), Positives = 64/125 (51%)
Frame = -1
Query: 152 RLDERRVLLDDLTEDDVIWTPYEAHREWRQRDERALFSGYIQCPYPPAVRPHLPERVMRQ 211
R+ R LD L DV W PY HRE + R+ +SG + C + P V + ERV+RQ
Sbjct: 409 RMPSYRERLDRLRIPDVCWIPYGEHREVQDFHVRSCYSGLL-C-WGPVVVYYRSERVVRQ 236
Query: 212 FGYIQTIPRHPSEMDRSSAAEVVDAAFADYVSYLFPKGDPVIEEGQAVGGYMDWYAIVSH 271
FGY QTIP S +D + + + + Y ++ P G+ + G Y+DW+ SH
Sbjct: 235 FGYTQTIP--ASTVDSWVSYDDIHDRWMHYEDHIVPAGEVCVVPGACSSDYIDWFFRTSH 62
Query: 272 CFIIP 276
F+ P
Sbjct: 61 PFMTP 47
>AI495080
Length = 407
Score = 71.6 bits (174), Expect = 5e-13
Identities = 43/122 (35%), Positives = 60/122 (48%), Gaps = 2/122 (1%)
Frame = +2
Query: 103 KQMGGYTSLLLAWVFEHFPDRLVRRYVSPDYIEDQARACRWKESRS--GHARLDERRVLL 160
+Q+ GY +LL W++EHFP + DY E RACRW ++ R R L
Sbjct: 2 RQLSGYITLLQCWIYEHFPS-VAESTAD*DYDEASPRACRWIATKKTVKSIRTPSYRERL 178
Query: 161 DDLTEDDVIWTPYEAHREWRQRDERALFSGYIQCPYPPAVRPHLPERVMRQFGYIQTIPR 220
+ L DV W Y HRE + R+ +SG + + + PE+V+RQFGY IP
Sbjct: 179 NRLRISDVCWILYGEHREVQDFHVRSCYSGLLH--WGLVAVYYRPEKVVRQFGYTPAIPA 352
Query: 221 HP 222
P
Sbjct: 353 PP 358
>TC211858
Length = 524
Score = 40.8 bits (94), Expect(3) = 6e-13
Identities = 17/46 (36%), Positives = 31/46 (66%)
Frame = -1
Query: 67 WIDMFQNLERVSEFAWGAMALATLYDQLGQSLRSGVKQMGGYTSLL 112
++D F++L + +AWG AL +Y+QL ++ R+ +Q+ GY +LL
Sbjct: 458 YLDAFRDLGQSGGYAWGVAALVHMYNQLDEASRTTTRQIVGYLTLL 321
Score = 40.8 bits (94), Expect(3) = 6e-13
Identities = 28/75 (37%), Positives = 36/75 (47%), Gaps = 5/75 (6%)
Frame = -3
Query: 108 YTSLLL--AWVFEHFPDRLVRRYVSPD-YIEDQARACRWKESRSGHARLDER--RVLLDD 162
Y +L+ W++EHFP V + V D Y E RA RW S++ + R D
Sbjct: 258 YVNLIFL*CWIYEHFPS--VHQCVIDDTY*ETSPRASRWLTSKAHMKGITGAPYRARCDA 85
Query: 163 LTEDDVIWTPYEAHR 177
LT DV W PY HR
Sbjct: 84 LTVIDVSWLPYTEHR 40
Score = 29.3 bits (64), Expect(3) = 6e-13
Identities = 12/15 (80%), Positives = 14/15 (93%)
Frame = -2
Query: 44 YLMHLVGATLFADKS 58
YL+HLVG TLFA+KS
Sbjct: 523 YLLHLVGCTLFANKS 479
>TC222177 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein, partial
(17%)
Length = 394
Score = 50.4 bits (119), Expect = 1e-06
Identities = 26/44 (59%), Positives = 27/44 (61%)
Frame = -3
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
A T G GRGG GA GRAG R+G R GGAG GGR RGG
Sbjct: 257 ARTSGGRRGGGRGGSGARGRAGTRNGARARGGAGVGGGRGARGG 126
Score = 30.4 bits (67), Expect = 1.4
Identities = 18/32 (56%), Positives = 18/32 (56%), Gaps = 3/32 (9%)
Frame = -3
Query: 347 AGGRSGGREGGGAGAMG--GRRG-RGGRRGRG 375
A SGGR GGG G G GR G R G R RG
Sbjct: 260 AARTSGGRRGGGRGGSGARGRAGTRNGARARG 165
>TC206524 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 464
Score = 49.3 bits (116), Expect = 3e-06
Identities = 25/38 (65%), Positives = 26/38 (67%), Gaps = 1/38 (2%)
Frame = +3
Query: 337 GRGGLGAGGRA-GGRSGGREGGGAGAMGGRRGRGGRRG 373
G GG GAGGR+ GGR GGR GGG G G RG GGR G
Sbjct: 12 GSGGRGAGGRSGGGRFGGRSGGGGGGRFGGRGGGGRFG 125
Score = 43.1 bits (100), Expect = 2e-04
Identities = 25/50 (50%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRR--GRG 375
G G GR G G GGR GGR GG GG+G G G GG R GRG
Sbjct: 36 GRSGGGRFGGRSGGGGGGRFGGRGGGGRFGGSGGGGRFGGSGGGRFGGRG 185
Score = 39.3 bits (90), Expect = 0.003
Identities = 25/50 (50%), Positives = 28/50 (56%)
Frame = +3
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
+G G GRGG G G +GG GGR GG G G RG GG RGRG+
Sbjct: 69 SGGGGGGRFGGRGGGGRFGGSGG--GGRFGGSGGGRFGGRG-GGGRGRGN 209
Score = 38.9 bits (89), Expect = 0.004
Identities = 20/33 (60%), Positives = 20/33 (60%)
Frame = +3
Query: 339 GGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGR 371
GG G GGR GG GGR GG GG RGRG R
Sbjct: 123 GGSGGGGRFGGSGGGRFGGRG---GGGRGRGNR 212
Score = 38.1 bits (87), Expect = 0.007
Identities = 23/41 (56%), Positives = 24/41 (58%), Gaps = 2/41 (4%)
Frame = +3
Query: 335 AAGRGGLGA-GGRAGGRSGGREGG-GAGAMGGRRGRGGRRG 373
A GR G G GGR+GG GGR GG G G G G GGR G
Sbjct: 30 AGGRSGGGRFGGRSGGGGGGRFGGRGGGGRFGGSGGGGRFG 152
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 48.9 bits (115), Expect = 4e-06
Identities = 25/48 (52%), Positives = 25/48 (52%)
Frame = +2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GG GG GGG G GG GRGGRR G
Sbjct: 254 GGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREEG 397
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/48 (50%), Positives = 25/48 (52%)
Frame = +2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GGR GG GGG G GG G+GG G G
Sbjct: 35 GGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGG 178
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/48 (50%), Positives = 25/48 (52%)
Frame = +2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GG GGR GGG G GG G GG +G G
Sbjct: 23 GGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGG 166
Score = 45.1 bits (105), Expect = 5e-05
Identities = 22/38 (57%), Positives = 22/38 (57%)
Frame = +2
Query: 338 RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
RGG G GG GG GG GGG G GGR G GG G G
Sbjct: 20 RGGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGG 133
Score = 45.1 bits (105), Expect = 5e-05
Identities = 24/48 (50%), Positives = 24/48 (50%)
Frame = +1
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GRGG G GG GG GG GGG G GG G GG G G
Sbjct: 61 GGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGG 204
Score = 44.7 bits (104), Expect = 7e-05
Identities = 24/48 (50%), Positives = 24/48 (50%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GG GG GGG G GGR G GG G G
Sbjct: 45 GGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGG 188
Score = 43.9 bits (102), Expect = 1e-04
Identities = 23/46 (50%), Positives = 23/46 (50%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G G G GG G GG GG GGR GGG G GG G GG G
Sbjct: 78 GGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGG 215
Score = 43.5 bits (101), Expect = 2e-04
Identities = 23/48 (47%), Positives = 23/48 (47%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG G GG GGG G GG GGRRG G
Sbjct: 261 GGGGGGGGGGGGGXGGGGGGRGEGGGGGGGGGGGGGGGGEGGGRRGGG 404
Score = 43.5 bits (101), Expect = 2e-04
Identities = 24/50 (48%), Positives = 25/50 (50%), Gaps = 2/50 (4%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRR--GRGGRRGRG 375
G G G GG G GG GG GG GGG G +GGR G GG G G
Sbjct: 117 GGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGG 266
Score = 43.1 bits (100), Expect = 2e-04
Identities = 22/46 (47%), Positives = 23/46 (49%)
Frame = +1
Query: 330 QGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
+G G GG G GG GG GG GGG G GG G GG G G
Sbjct: 241 EGGERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGGGGGGGGGEG 378
Score = 42.7 bits (99), Expect = 3e-04
Identities = 23/47 (48%), Positives = 24/47 (50%)
Frame = +1
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGR 374
G +G G GG G GG GG GG GGG G GG G GG GR
Sbjct: 247 GERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGGGG--GGGGGEGR 381
Score = 42.7 bits (99), Expect = 3e-04
Identities = 23/48 (47%), Positives = 23/48 (47%)
Frame = +1
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GGR G GGG G GG G GG G G
Sbjct: 25 GGGGGGGGGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGG 168
Score = 42.7 bits (99), Expect = 3e-04
Identities = 22/39 (56%), Positives = 22/39 (56%)
Frame = +2
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GRGG G GG GG GG GGG G GG G GG G G
Sbjct: 239 GRGGRGGGGGGGGGGGG--GGGGGGGGGGEGGGGGGGGG 349
Score = 42.4 bits (98), Expect = 3e-04
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 8/56 (14%)
Frame = +2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRG--------GRRGRG 375
G G G GG G GG GG+ GG GGG G GG G G GR GRG
Sbjct: 89 GGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRG 256
Score = 42.0 bits (97), Expect = 5e-04
Identities = 21/39 (53%), Positives = 21/39 (53%)
Frame = +3
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GRG G GG GG GG GGG G GG G GG G G
Sbjct: 6 GRGLPGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGG 122
Score = 42.0 bits (97), Expect = 5e-04
Identities = 23/48 (47%), Positives = 23/48 (47%)
Frame = +2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GG GG GGG G GGR GR G G
Sbjct: 263 GGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREEGGGG 406
Score = 41.2 bits (95), Expect = 8e-04
Identities = 24/56 (42%), Positives = 25/56 (43%), Gaps = 8/56 (14%)
Frame = +1
Query: 328 GTQGIAYAAGRGGLGAGGRAGG--------RSGGREGGGAGAMGGRRGRGGRRGRG 375
G +G G GG G GG GG R GG GGG G GG G GG G G
Sbjct: 148 GGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGGERGGGGGGGGGGGGGGGXGGGG 315
Score = 39.3 bits (90), Expect = 0.003
Identities = 22/48 (45%), Positives = 23/48 (47%)
Frame = +2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G + G G GGR GG GG GGG G GG G GG G G
Sbjct: 197 GGGGGGFFRGERXXGRGGRGGGGGGG--GGGGGGGGGGGGGGGEGGGG 334
Score = 38.5 bits (88), Expect = 0.005
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 338 RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
R G GG GG GG GGG G GG GRGG G G
Sbjct: 238 REGGERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGG 351
Score = 38.1 bits (87), Expect = 0.007
Identities = 22/49 (44%), Positives = 23/49 (46%), Gaps = 1/49 (2%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGG-REGGGAGAMGGRRGRGGRRGRG 375
G G + GR G GG GG GG GGG G GG GRG G G
Sbjct: 198 GGGGGGFLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGGGRGEGGGGG 344
Score = 38.1 bits (87), Expect = 0.007
Identities = 26/61 (42%), Positives = 27/61 (43%), Gaps = 13/61 (21%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGRE--------G-----GGAGAMGGRRGRGGRRGR 374
G +G G GG G GG GG GGRE G GG G GG G GG RG
Sbjct: 150 GGRGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGGGRGE 329
Query: 375 G 375
G
Sbjct: 330 G 332
Score = 30.4 bits (67), Expect = 1.4
Identities = 16/32 (50%), Positives = 16/32 (50%)
Frame = +1
Query: 344 GGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GG GG GGG G GG G GG GRG
Sbjct: 1 GGGEVCPGGGGGGGGGGGGGGGGGGGGGGGRG 96
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 46.6 bits (109), Expect = 2e-05
Identities = 25/52 (48%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Frame = -3
Query: 328 GTQGIAYAAGRGGLGAGG----RAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G+QG +G GG G GG GG SGG GGG G GG G GG +G G
Sbjct: 460 GSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGGGGGGYGGGGDKGGG 305
Score = 43.1 bits (100), Expect = 2e-04
Identities = 22/46 (47%), Positives = 24/46 (51%)
Frame = -3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G+ G G G G GG GG SGG EGGG+ GG G GG G
Sbjct: 475 GSGGGGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGGGGG 338
Score = 41.6 bits (96), Expect = 6e-04
Identities = 24/52 (46%), Positives = 31/52 (59%), Gaps = 3/52 (5%)
Frame = -3
Query: 328 GTQGIAYAAGRGGL--GAGGRAGGRSG-GREGGGAGAMGGRRGRGGRRGRGH 376
G+QG +AG GG G+GG AGG G G +GGG+G G + G G G G+
Sbjct: 574 GSQGGGGSAGGGGS*GGSGGSAGGGEGSGSQGGGSGGGGSQGGGSGSGGGGY 419
Score = 39.7 bits (91), Expect = 0.002
Identities = 21/48 (43%), Positives = 25/48 (51%), Gaps = 1/48 (2%)
Frame = -3
Query: 327 AGTQGIAYAAG-RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
+G+ G Y G GG GG +GG GG GGG G GG +GG G
Sbjct: 442 SGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGGGGGGYGGGGDKGGGIG 299
Score = 39.3 bits (90), Expect = 0.003
Identities = 24/55 (43%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Frame = -3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGR-----SGGREGG--GAGAMGGRRGRGGRRGRG 375
G++G G G G GG AGG SGG GG G+G+ GG G GG +G G
Sbjct: 607 GSKGGGGYGGGGSQGGGGSAGGGGS*GGSGGSAGGGEGSGSQGGGSGGGGSQGGG 443
Score = 35.4 bits (80), Expect = 0.042
Identities = 21/48 (43%), Positives = 24/48 (49%)
Frame = -3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G+ G +G G G+GG GG GG G G G GG G GG G G
Sbjct: 517 GSAGGGEGSGSQGGGSGG--GGSQGGGSGSGGGGYGG-GGSGGSEGGG 383
Score = 32.7 bits (73), Expect = 0.27
Identities = 18/37 (48%), Positives = 19/37 (50%)
Frame = -3
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G G G GG GG GG GGG+ GG G GG G
Sbjct: 631 GGGSAGGGGSKGG--GGYGGGGSQGGGGSAGGGGS*G 527
Score = 32.0 bits (71), Expect = 0.47
Identities = 22/51 (43%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Frame = -3
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGG--AGAMGGRRGRGGRRGRG 375
+G+QG GG G GG GG SG GGG G GG G G G G
Sbjct: 493 SGSQG-------GGSGGGGSQGGGSGSG-GGGYGGGGSGGSEGGGSSGGGG 365
>TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protein [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(88%)
Length = 1085
Score = 46.2 bits (108), Expect = 2e-05
Identities = 24/50 (48%), Positives = 29/50 (58%)
Frame = +2
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
AG++ +YA R G G+ GR+ GR GG GGG G GG G G G GH
Sbjct: 647 AGSEAGSYAGSRAGSGS-GRSQGRGGGGGGGGGGGGGGGSGYGHGEGYGH 793
Score = 42.4 bits (98), Expect = 3e-04
Identities = 20/41 (48%), Positives = 24/41 (57%)
Frame = +2
Query: 335 AAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
++G GG GAG AG +G R G G+G GR G GG G G
Sbjct: 623 SSGSGGSGAGSEAGSYAGSRAGSGSGRSQGRGGGGGGGGGG 745
Score = 28.1 bits (61), Expect = 6.8
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 1/35 (2%)
Frame = +2
Query: 331 GIAYAAGRG-GLGAGGRAGGRSGGREGGGAGAMGG 364
G+ G G GLG GG +G +G G G+G+ G
Sbjct: 476 GLGVGLGLGLGLGIGGGSGSGAGAGAGSGSGSGSG 580
>BM954311 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (9%)
Length = 406
Score = 45.8 bits (107), Expect = 3e-05
Identities = 22/38 (57%), Positives = 24/38 (62%)
Frame = -2
Query: 334 YAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGR 371
+ G GG GAGGR GGR+G GGG G MG G GGR
Sbjct: 381 FGEGEGGSGAGGRGGGRTGRGGGGGDG*MGRGGGGGGR 268
Score = 27.7 bits (60), Expect = 8.8
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 3/39 (7%)
Frame = -2
Query: 339 GGLGAGGRAGGRSGG---REGGGAGAMGGRRGRGGRRGR 374
GG G G + G GG EGGG G G GG+ G+
Sbjct: 183 GGGGGGLKCEGGGGG*KDCEGGGGGLKGWEGDGGGKVGK 67
>BQ576456
Length = 424
Score = 45.1 bits (105), Expect = 5e-05
Identities = 27/73 (36%), Positives = 39/73 (52%)
Frame = +1
Query: 19 MRFSFLRDLYPTAVRERRYEHAARMYLMHLVGATLFADKSGGHSFSARWIDMFQNLERVS 78
+R S+LR++Y + + R + A R YL HLV TLFA+KS H + F +L
Sbjct: 208 VRLSWLREVYQSRCQARCWIVAVRAYLFHLVDCTLFANKSATHVHVVH-LKGF*DLC*SG 384
Query: 79 EFAWGAMALATLY 91
+ WG AL +Y
Sbjct: 385 GYGWGVAALVHMY 423
>TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (66%)
Length = 773
Score = 44.7 bits (104), Expect = 7e-05
Identities = 20/39 (51%), Positives = 23/39 (58%)
Frame = +1
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRR 366
G +G + A GG G GGR GGR GGR GG G GG +
Sbjct: 127 GDRGTPFKARGGGRGGGGRGGGRGGGRGGGRGGMKGGSK 243
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/46 (54%), Positives = 26/46 (56%), Gaps = 9/46 (19%)
Frame = +1
Query: 337 GRGGLGAGG--------RAGGRSGGREGGGAGAMGGR-RGRGGRRG 373
GRGG G GG R GGR GG GGG G GGR GRGG +G
Sbjct: 103 GRGGGGRGGDRGTPFKARGGGRGGGGRGGGRG--GGRGGGRGGMKG 234
Score = 39.7 bits (91), Expect = 0.002
Identities = 26/50 (52%), Positives = 26/50 (52%), Gaps = 13/50 (26%)
Frame = +1
Query: 337 GRGGLGAGG--RAGGRSG---GREGGGAG--------AMGGRRGRGGRRG 373
GRGG G GG R GGR GR GGG G A GG RG GGR G
Sbjct: 40 GRGGFGGGGGFRGGGRGDRGRGRGGGGRGGDRGTPFKARGGGRGGGGRGG 189
Score = 38.1 bits (87), Expect = 0.007
Identities = 22/45 (48%), Positives = 22/45 (48%), Gaps = 6/45 (13%)
Frame = +1
Query: 337 GRGGLGAGGRAGGRSGGR------EGGGAGAMGGRRGRGGRRGRG 375
GRG G G GGR G R GGG G G GRGG RG G
Sbjct: 82 GRGDRGRGRGGGGRGGDRGTPFKARGGGRGGGGRGGGRGGGRGGG 216
>TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wall structural
protein 1.0 precursor (GRP 1.0), partial (27%)
Length = 447
Score = 43.9 bits (102), Expect = 1e-04
Identities = 24/49 (48%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Frame = +3
Query: 328 GTQGIAYAAGRG-GLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G Y G G G GAGG GG + G GGG G GG RGG G G
Sbjct: 21 GAHGGGYGGGEGAGQGAGGGYGGGAVGGGGGGGGGSGGGGARGGGYGAG 167
Score = 42.4 bits (98), Expect = 3e-04
Identities = 25/50 (50%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Frame = +3
Query: 328 GTQGIAYAAGRGG-LGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G A+ G GG GAG AGG GG GG G GG G GG RG G+
Sbjct: 9 GGAGGAHGGGYGGGEGAGQGAGGGYGGGAVGGGGGGGGGSGGGGARGGGY 158
Score = 38.1 bits (87), Expect = 0.007
Identities = 23/46 (50%), Positives = 25/46 (54%)
Frame = +3
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G A GG G GG +GG GG GGG GA GG G GG G G+
Sbjct: 78 GYGGGAVGGGGGGGGGSGG--GGARGGGYGA-GGGSGEGGGHGGGY 206
Score = 33.1 bits (74), Expect = 0.21
Identities = 19/41 (46%), Positives = 20/41 (48%), Gaps = 1/41 (2%)
Frame = +3
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGR-EGGGAGAMGGRRG 367
G G A G GG G G G R GG GGG+G GG G
Sbjct: 78 GYGGGAVGGGGGGGGGSGGGGARGGGYGAGGGSGEGGGHGG 200
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 42.4 bits (98), Expect = 3e-04
Identities = 21/39 (53%), Positives = 21/39 (53%)
Frame = +1
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GG G GG GGR GG GGG G GG GG G G
Sbjct: 337 GGGGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYG 453
Score = 41.2 bits (95), Expect = 8e-04
Identities = 26/57 (45%), Positives = 27/57 (46%), Gaps = 8/57 (14%)
Frame = +1
Query: 328 GTQGIAYAAGRGG-LGAGGRAGGRS-------GGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y GRGG G GGR GG GG GGG GG G GG R RG+
Sbjct: 346 GGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYGGGGGYGGGGGGGYGGGRDRGY 516
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/41 (53%), Positives = 22/41 (53%)
Frame = +1
Query: 335 AAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
A RGG G GG GG GG GGG G GGR G GG G
Sbjct: 316 AQSRGGGGGGGGGGGGYGGGRGGGYGG-GGRGGGGGYNRSG 435
Score = 33.5 bits (75), Expect = 0.16
Identities = 22/41 (53%), Positives = 23/41 (55%), Gaps = 2/41 (4%)
Frame = +1
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRG--RGGRRGRG 375
G GG G GG GG GGR+ G G GG RG RGG G G
Sbjct: 454 GGGGYGGGG-GGGYGGGRDRGYGG--GGDRGYSRGGDGGDG 567
>TC234144
Length = 468
Score = 42.4 bits (98), Expect = 3e-04
Identities = 19/38 (50%), Positives = 24/38 (63%)
Frame = +3
Query: 157 RVLLDDLTEDDVIWTPYEAHREWRQRDERALFSGYIQC 194
R LD L D++W PY HRE R + +LFSGYI+C
Sbjct: 6 RPALDML*TKDILWMPYTVHREHRPFHDISLFSGYIRC 119
>TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protein, partial
(50%)
Length = 434
Score = 42.4 bits (98), Expect = 3e-04
Identities = 26/62 (41%), Positives = 32/62 (50%), Gaps = 3/62 (4%)
Frame = +3
Query: 317 KRALRILQDLAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRG---RGGRRG 373
++ L + + L +G + G GG G GG GGR GG G G GGR G GG RG
Sbjct: 177 RKLLPLARFLPQPKGQSAGRGGGGGGRGGGRGGRGGGGFRGRGGPRGGRGGPPRGGGFRG 356
Query: 374 RG 375
RG
Sbjct: 357 RG 362
>TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (clone
na-481-5), partial (20%)
Length = 752
Score = 42.0 bits (97), Expect = 5e-04
Identities = 24/39 (61%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Frame = +2
Query: 337 GRGGLGAGGRAGGRSGGREG--GGAGAMGGRRGRGGRRG 373
G GG GAGGR+GG GGR G GG G G RG GGR G
Sbjct: 275 GSGGRGAGGRSGG--GGRFGARGGGGGRFGARGGGGRFG 385
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/45 (53%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Frame = +2
Query: 328 GTQGIAYAAGR-GGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGR 371
G G GR GG G GGR GG GGR GG GG RGRG R
Sbjct: 347 GRFGARGGGGRFGGSGGGGRFGGSGGGRFGGRG---GGGRGRGNR 472
Score = 37.7 bits (86), Expect = 0.009
Identities = 27/64 (42%), Positives = 32/64 (49%), Gaps = 14/64 (21%)
Frame = +2
Query: 327 AGTQGIAYAAGRGG-LGAGGRAGGRSGGREGGG-----------AGAMGGRRG--RGGRR 372
+G +G +G GG GA G GGR G R GGG G+ GGR G GG R
Sbjct: 278 SGGRGAGGRSGGGGRFGARGGGGGRFGARGGGGRFGGSGGGGRFGGSGGGRFGGRGGGGR 457
Query: 373 GRGH 376
GRG+
Sbjct: 458 GRGN 469
Score = 35.8 bits (81), Expect = 0.032
Identities = 22/43 (51%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Frame = +2
Query: 335 AAGRGGLGAGGRAGGRSGGREGGGA--GAMGGRRGRGGRRGRG 375
A R G+GGR +GGR GGG GA GG GR G RG G
Sbjct: 254 AKPRDNQGSGGRG---AGGRSGGGGRFGARGGGGGRFGARGGG 373
>BE475635
Length = 412
Score = 42.0 bits (97), Expect = 5e-04
Identities = 21/61 (34%), Positives = 37/61 (60%)
Frame = +3
Query: 48 LVGATLFADKSGGHSFSARWIDMFQNLERVSEFAWGAMALATLYDQLGQSLRSGVKQMGG 107
L+G TLFA+KS H + ++D F++L + +A +AL +YD L + +S +++ G
Sbjct: 3 LLGCTLFANKSATHVYVI-FLDAFRDLSQSGSYA*VVVALVHMYDHLNDACKSDDRKLAG 179
Query: 108 Y 108
Y
Sbjct: 180 Y 182
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 41.6 bits (96), Expect = 6e-04
Identities = 23/44 (52%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Frame = +3
Query: 337 GRGGLGAGGRA-----GGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GRGG G GGR GG GG GGG G GG G GG G G
Sbjct: 99 GRGGYGGGGRGGRGGYGGGGGGYGGGGYGGGGGYGGGGGGGGGG 230
Score = 36.2 bits (82), Expect = 0.025
Identities = 20/49 (40%), Positives = 22/49 (44%)
Frame = +3
Query: 325 DLAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
D+ G RGG G + G GGR G G G GGR G GG G
Sbjct: 15 DVXGPDESPVQGTRGGGGXXRGSYGXGGGRGGYGGGGRGGRGGYGGGGG 161
>BU927072
Length = 314
Score = 41.6 bits (96), Expect = 6e-04
Identities = 22/36 (61%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Frame = -2
Query: 339 GGLGAGGRAGGR-SGGREGGGAGAMGGRRGRGGRRG 373
G G GGR GG GGR GGGAG GG R R GR G
Sbjct: 304 GRXGGGGRTGGGVGGGRGGGGAGGGGGGRRRRGRSG 197
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/45 (48%), Positives = 23/45 (50%)
Frame = -2
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G+ G GG GG GGR GG GGG G RRGR G R G
Sbjct: 313 GVGGRXGGGGRTGGGVGGGRGGGGAGGGGGGR-RRRGRSGFRNGG 182
Score = 40.4 bits (93), Expect = 0.001
Identities = 21/36 (58%), Positives = 22/36 (60%), Gaps = 2/36 (5%)
Frame = -2
Query: 342 GAGGRAGG--RSGGREGGGAGAMGGRRGRGGRRGRG 375
G GGR GG R+GG GGG G G G GGRR RG
Sbjct: 313 GVGGRXGGGGRTGGGVGGGRGGGGAGGGGGGRRRRG 206
Score = 39.7 bits (91), Expect = 0.002
Identities = 26/52 (50%), Positives = 27/52 (51%), Gaps = 5/52 (9%)
Frame = -2
Query: 328 GTQGIAYAAGRGGLGAGGRAGGR-----SGGREGGGAGAMGGRRGRGGRRGR 374
G G GRGG GAGG GGR SG R GGG+ G RRG G GR
Sbjct: 286 GRTGGGVGGGRGGGGAGGGGGGRRRRGRSGFRNGGGS---GWRRGGGWWSGR 140
Score = 33.1 bits (74), Expect = 0.21
Identities = 18/39 (46%), Positives = 18/39 (46%)
Frame = -2
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GG GG AGG GGR G G G RRG G
Sbjct: 271 GVGGGRGGGGAGGGGGGRRRRGRSGFRNGGGSGWRRGGG 155
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,006,053
Number of Sequences: 63676
Number of extensions: 229579
Number of successful extensions: 4525
Number of sequences better than 10.0: 441
Number of HSP's better than 10.0 without gapping: 2875
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3898
length of query: 376
length of database: 12,639,632
effective HSP length: 99
effective length of query: 277
effective length of database: 6,335,708
effective search space: 1754991116
effective search space used: 1754991116
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0348.16


