

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0347b.6
(1014 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
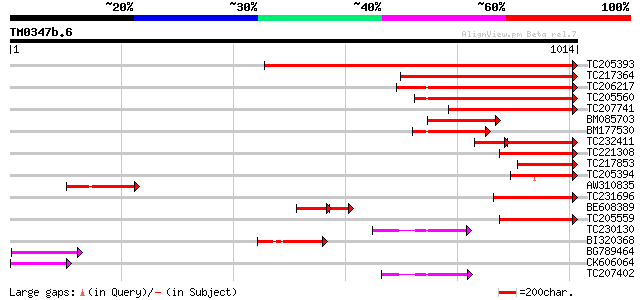
Score E
Sequences producing significant alignments: (bits) Value
TC205393 UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase, partial... 934 0.0
TC217364 plasma membrane Ca2+-ATPase 511 e-145
TC206217 similar to UP|ACA9_ARATH (Q9LU41) Potential calcium-tra... 363 e-100
TC205560 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transpor... 343 2e-94
TC207741 similar to UP|Q8W0V0 (Q8W0V0) Type IIB calcium ATPase, ... 310 2e-84
BM085703 homologue to SP|Q9LIK7|ACA Potential calcium-transporti... 202 6e-52
BM177530 homologue to GP|16508164|g type IIB calcium ATPase {Med... 200 2e-51
TC232411 similar to UP|Q8W0V0 (Q8W0V0) Type IIB calcium ATPase, ... 134 3e-51
TC221308 similar to UP|Q9FVE7 (Q9FVE7) Plasma membrane Ca2+-ATPa... 181 2e-45
TC217853 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-AT... 167 2e-41
TC205394 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-AT... 153 3e-37
AW310835 similar to GP|16508162|g type IIB calcium ATPase {Medic... 135 9e-32
TC231696 weakly similar to UP|ACAA_ARATH (Q9SZR1) Potential calc... 134 3e-31
BE608389 homologue to GP|11066056|g plasma membrane Ca2+-ATPase ... 87 2e-29
TC205559 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transpor... 126 4e-29
TC230130 similar to UP|AHM7_ARATH (Q9SH30) Potential copper-tran... 98 2e-20
BI320368 similar to SP|Q9LU41|ACA Potential calcium-transporting... 97 5e-20
BG789464 similar to GP|16508162|g type IIB calcium ATPase {Medic... 91 3e-18
CK606064 90 4e-18
TC207402 similar to UP|Q941L1 (Q941L1) Copper-transporting P-typ... 79 1e-14
>TC205393 UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase, partial (55%)
Length = 2009
Score = 934 bits (2415), Expect = 0.0
Identities = 467/560 (83%), Positives = 517/560 (91%), Gaps = 1/560 (0%)
Frame = +2
Query: 456 KTGTLTTNHMTVVKTCICMSSKEV-NNKEHGLCSELPDSAQKLLLQSIFNNTGGEVVVNK 514
KTGTLTTNHMTVVKTC CM+SKEV NN LCSELP+ A KLLL+SIFNNTGGEVVVN+
Sbjct: 2 KTGTLTTNHMTVVKTCFCMNSKEVSNNNASSLCSELPEPAVKLLLESIFNNTGGEVVVNQ 181
Query: 515 RGKREILGTPTESAILEFGLSLGGDPQKERQACKLVKVEPFNSQKKRMGVVVELPEGGLR 574
GKREILGTPTE+AILEFGLSLGGD Q E+QACKLVKVEPFNS KK+M VVVELP GGLR
Sbjct: 182 NGKREILGTPTEAAILEFGLSLGGDFQGEKQACKLVKVEPFNSTKKKMSVVVELPGGGLR 361
Query: 575 AHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHG 634
AHCKGASEI+LAACD V++S G+VVPL+ ES ++L++TI+QFA EALRTLCLAY+ELE+G
Sbjct: 362 AHCKGASEIILAACDKVLNSNGEVVPLDEESTSHLKATINQFASEALRTLCLAYVELENG 541
Query: 635 FSAEDPIPASGYTCIGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIAREC 694
FS EDPIP SGYTCIGV+GIKDPVRPGVKESV + RSAGI VRMVTGDNINTAKAIAREC
Sbjct: 542 FSPEDPIPVSGYTCIGVIGIKDPVRPGVKESVAM*RSAGITVRMVTGDNINTAKAIAREC 721
Query: 695 GILTEDGLAIEGPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTG 754
GILT+DG+AIEGP+FREK+Q+E+ ELIPKIQVMARSSPLDKHTLVK LRTTFGEVVAVTG
Sbjct: 722 GILTDDGIAIEGPEFREKSQKELLELIPKIQVMARSSPLDKHTLVKHLRTTFGEVVAVTG 901
Query: 755 DGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFV 814
DGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFV
Sbjct: 902 DGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFV 1081
Query: 815 QFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAP 874
QFQLTVNVVAL+VNF+SA LTG+APLTAVQLLWVNMIMDTLGALALATEPP DDLMKR+P
Sbjct: 1082 QFQLTVNVVALIVNFTSACLTGTAPLTAVQLLWVNMIMDTLGALALATEPPNDDLMKRSP 1261
Query: 875 LGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFC 934
+GRKG+FI+++MWRNILGQ+LYQF+VIWFLQ+ GK +F L GPN+ +VLNTLIFNSFVFC
Sbjct: 1262 VGRKGNFISNVMWRNILGQSLYQFMVIWFLQSRGKSIFLLEGPNSDLVLNTLIFNSFVFC 1441
Query: 935 QVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIF 994
QVFNEINSREME+++VFKGI DN+VFV VI TV FQIIIVEYLGTFANTTPL+L QW F
Sbjct: 1442 QVFNEINSREMEKINVFKGILDNYVFVGVISATVFFQIIIVEYLGTFANTTPLTLSQWFF 1621
Query: 995 CLSVGYVGMPIATYLKQIPV 1014
CL VG++GMPIA LK+IPV
Sbjct: 1622 CLLVGFMGMPIAARLKKIPV 1681
>TC217364 plasma membrane Ca2+-ATPase
Length = 1375
Score = 511 bits (1315), Expect = e-145
Identities = 254/315 (80%), Positives = 288/315 (90%)
Frame = +3
Query: 700 DGLAIEGPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTND 759
DG+AIEGP+FREK++EE+ ++IPKIQVMARSSP+DKHTLVK LRTTF EVV+VTGDGTND
Sbjct: 6 DGIAIEGPEFREKSEEELLDIIPKIQVMARSSPMDKHTLVKHLRTTFQEVVSVTGDGTND 185
Query: 760 APALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLT 819
APALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLT
Sbjct: 186 APALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLT 365
Query: 820 VNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKG 879
VNVVAL+VNFSSA LTG+APLTAVQLLWVNMIMDTLGALALATEPP D+LMKR P+GRKG
Sbjct: 366 VNVVALIVNFSSACLTGNAPLTAVQLLWVNMIMDTLGALALATEPPNDELMKRPPVGRKG 545
Query: 880 DFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQVFNE 939
+FI+++MWRNILGQ++YQFVVIWFLQT GK F L GP++ ++LNTLIFNSFVFCQVFNE
Sbjct: 546 NFISNVMWRNILGQSIYQFVVIWFLQTRGKVTFHLDGPDSDLILNTLIFNSFVFCQVFNE 725
Query: 940 INSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVG 999
I+SR+ME ++VF+GI N+VFVAV+ TVVFQIIIVE+LGTFANT+PLSL QW + G
Sbjct: 726 ISSRDMERINVFEGILKNYVFVAVLTSTVVFQIIIVEFLGTFANTSPLSLKQWFGSVLFG 905
Query: 1000 YVGMPIATYLKQIPV 1014
+GMPIA LK IPV
Sbjct: 906 VLGMPIAAALKMIPV 950
>TC206217 similar to UP|ACA9_ARATH (Q9LU41) Potential calcium-transporting
ATPase 9, plasma membrane-type (Ca(2+)-ATPase isoform 9)
, partial (32%)
Length = 1383
Score = 363 bits (931), Expect = e-100
Identities = 189/329 (57%), Positives = 241/329 (72%), Gaps = 7/329 (2%)
Frame = +1
Query: 693 ECGILT--EDGLA---IEGPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFG 747
ECGIL ED + IEG FRE +++E ++ KI VM RSSP DK LV+ LR G
Sbjct: 4 ECGILASIEDAVEPNIIEGKKFRELSEKEREDIAKKITVMGRSSPNDKLLLVQALRKG-G 180
Query: 748 EVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVY 807
EVVAVTGDGTNDAPALHEADIGL+MGI GTEVAKES+D+IILDDNF+++V V +WGRSVY
Sbjct: 181 EVVAVTGDGTNDAPALHEADIGLSMGIQGTEVAKESSDIIILDDNFASVVKVVRWGRSVY 360
Query: 808 INIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTD 867
NIQKF+QFQLTVNV AL++N +A+ +G PL AVQLLWVN+IMDTLGALALATEPPTD
Sbjct: 361 ANIQKFIQFQLTVNVAALVINVVAAITSGDVPLNAVQLLWVNLIMDTLGALALATEPPTD 540
Query: 868 DLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGV--VLNT 925
LM R+P+GR+ I +IMWRN++ QA+YQ V+ L G+ + + A V NT
Sbjct: 541 RLMHRSPVGRRESLITNIMWRNLIVQAVYQIAVLLVLNFCGESILPKQNTRADAFQVKNT 720
Query: 926 LIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTT 985
LIFN+FV CQ+FNE N+R+ +E++VF+G+ N +FV ++G T + QIII+E+LG F +T
Sbjct: 721 LIFNAFVLCQIFNEFNARKPDEMNVFRGVTKNKLFVGIVGVTFILQIIIIEFLGKFTSTV 900
Query: 986 PLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
L W+ L +G+V P+A K IPV
Sbjct: 901 RLDWKLWLASLGIGFVSWPLAIVGKFIPV 987
>TC205560 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transporting
ATPase-like protein {Arabidopsis thaliana;} , partial
(27%)
Length = 1356
Score = 343 bits (881), Expect = 2e-94
Identities = 178/293 (60%), Positives = 220/293 (74%), Gaps = 3/293 (1%)
Frame = +1
Query: 725 QVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESA 784
QVM RSSP DK LV+ LR G VVAVTGDGTNDAPALHEADIGLAMGI GTEVAKES+
Sbjct: 7 QVMGRSSPNDKLLLVQALRRK-GHVVAVTGDGTNDAPALHEADIGLAMGIQGTEVAKESS 183
Query: 785 DVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQ 844
D+IILDDNF+++V V +WGRSVY NIQKF+QFQLTVNV AL++N +AV +G PL AVQ
Sbjct: 184 DIIILDDNFASVVKVVRWGRSVYANIQKFIQFQLTVNVAALVINVVAAVSSGDVPLNAVQ 363
Query: 845 LLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFL 904
LLWVN+IMDTLGALALATEPPTD LM R P+GR+ I +IMWRN+L QA+YQ V+ L
Sbjct: 364 LLWVNLIMDTLGALALATEPPTDHLMDRTPVGRREPLITNIMWRNLLIQAMYQVSVLLVL 543
Query: 905 QTVGKWVFFL---RGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFV 961
G + L R +A V NTLIFN+FV CQ+FNE N+R+ +E ++FKG+ N++F+
Sbjct: 544 NFRGISILGLSHDRKDHAIKVKNTLIFNAFVLCQIFNEFNARKPDEFNIFKGVTRNYLFM 723
Query: 962 AVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
+IG TVV QI+I+ +LG F T L+ QW+ + +G +G P+A K IPV
Sbjct: 724 GIIGLTVVLQIVIILFLGKFTTTVRLNWKQWLISVVIGLIGWPLAVIGKLIPV 882
>TC207741 similar to UP|Q8W0V0 (Q8W0V0) Type IIB calcium ATPase, partial (24%)
Length = 947
Score = 310 bits (794), Expect = 2e-84
Identities = 151/230 (65%), Positives = 188/230 (81%)
Frame = +3
Query: 785 DVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQ 844
DVII+DDNF+TIV VA+WGR++YINIQKFVQFQLTVN+VAL++NF SA +TGSAPLTAVQ
Sbjct: 3 DVIIMDDNFTTIVNVARWGRAIYINIQKFVQFQLTVNIVALIINFVSACITGSAPLTAVQ 182
Query: 845 LLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFL 904
LLWVN+IMDTLGALALATEPP D LM R P+GR +FI MWRNI GQ+LYQ +V+ L
Sbjct: 183 LLWVNLIMDTLGALALATEPPNDGLMLRPPVGRTTNFITKPMWRNIFGQSLYQLIVLAVL 362
Query: 905 QTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVI 964
GK + + GP+A +VLNTLIFNSFVFCQVFNEINSRE+E++++FKG++++ +F VI
Sbjct: 363 TFDGKRLLRINGPDATIVLNTLIFNSFVFCQVFNEINSREIEKINIFKGMFESWIFFTVI 542
Query: 965 GCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
TVVFQ++IVE+LGTFA+T PLS W+ + +G MPI+ LK IPV
Sbjct: 543 FSTVVFQVLIVEFLGTFASTVPLSWQFWVLSVVIGAFSMPISVILKCIPV 692
>BM085703 homologue to SP|Q9LIK7|ACA Potential calcium-transporting ATPase 13
plasma membrane-type (EC 3.6.3.8) (Ca(2+)-ATPase
isoform, partial (12%)
Length = 411
Score = 202 bits (514), Expect = 6e-52
Identities = 99/131 (75%), Positives = 114/131 (86%)
Frame = +3
Query: 747 GEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSV 806
G VVAVTGDGTNDAPAL EADIGL+MGI GTEVAKES+D++ILDDNF+++VTV +WGR V
Sbjct: 6 GHVVAVTGDGTNDAPALKEADIGLSMGIQGTEVAKESSDIVILDDNFASVVTVLRWGRCV 185
Query: 807 YINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPT 866
Y NIQKF+QFQLTVNV AL +NF +AV G PLTAVQLLWVN+IMDTLGALALATE PT
Sbjct: 186 YNNIQKFIQFQLTVNVAALAINFVAAVSAGKVPLTAVQLLWVNLIMDTLGALALATEKPT 365
Query: 867 DDLMKRAPLGR 877
+LM + P+GR
Sbjct: 366 MELMHKPPVGR 398
>BM177530 homologue to GP|16508164|g type IIB calcium ATPase {Medicago
truncatula}, partial (13%)
Length = 422
Score = 200 bits (509), Expect = 2e-51
Identities = 101/140 (72%), Positives = 120/140 (85%)
Frame = +2
Query: 721 IPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVA 780
+ KI+VMARSSPLDK +V+ L+ G VVAVTGDGTNDAPAL EADIGL+MGI GTEVA
Sbjct: 5 VEKIRVMARSSPLDKLLMVQCLKKK-GHVVAVTGDGTNDAPALKEADIGLSMGIQGTEVA 181
Query: 781 KESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPL 840
KES+D++ILDDNF+++ TV +WGR VY NIQKF+QFQLTVNV AL++NF +AV +G PL
Sbjct: 182 KESSDIVILDDNFNSVATVLRWGRCVYNNIQKFIQFQLTVNVAALVINFVAAVSSGDVPL 361
Query: 841 TAVQLLWVNMIMDTLGALAL 860
T VQLLWVN+IMDTLGALAL
Sbjct: 362 TTVQLLWVNLIMDTLGALAL 421
>TC232411 similar to UP|Q8W0V0 (Q8W0V0) Type IIB calcium ATPase, partial (19%)
Length = 802
Score = 134 bits (336), Expect(2) = 3e-51
Identities = 62/124 (50%), Positives = 92/124 (74%)
Frame = +2
Query: 891 LGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDV 950
L Q++YQ +++ L GK + L G +A +LNTLIFNSFVFCQVFNEINSR+++++++
Sbjct: 197 LFQSIYQLIILGILNFDGKRLLGLSGSDATKILNTLIFNSFVFCQVFNEINSRDIDKINI 376
Query: 951 FKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLK 1010
F+G++D+ +F+A+I T FQ++IVE+LGTFA+T PL+ W+ + +G MPIA LK
Sbjct: 377 FRGMFDSWIFMAIIFATAAFQVVIVEFLGTFASTVPLNWQFWLLSVVIGAFSMPIAAILK 556
Query: 1011 QIPV 1014
IPV
Sbjct: 557 CIPV 568
Score = 87.8 bits (216), Expect(2) = 3e-51
Identities = 44/64 (68%), Positives = 50/64 (77%)
Frame = +3
Query: 831 SAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAPLGRKGDFINSIMWRNI 890
S L+ SAPLTAVQLLWVN+IMDTLGALALATEPP D L+KR P+ R +FI MWRNI
Sbjct: 18 SLSLSLSAPLTAVQLLWVNLIMDTLGALALATEPPNDGLLKRPPVARGANFITKPMWRNI 197
Query: 891 LGQA 894
+A
Sbjct: 198 FFKA 209
>TC221308 similar to UP|Q9FVE7 (Q9FVE7) Plasma membrane Ca2+-ATPase, partial
(14%)
Length = 790
Score = 181 bits (458), Expect = 2e-45
Identities = 89/138 (64%), Positives = 109/138 (78%)
Frame = +2
Query: 877 RKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQV 936
RK +FI+++MWRNI ++ V WFLQT GK F L GP++ ++LNTLIFNSFVFCQV
Sbjct: 2 RKENFISNVMWRNIXXXSIXXXVXXWFLQTRGKVXFHLDGPDSDLILNTLIFNSFVFCQV 181
Query: 937 FNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCL 996
FNEI+SR+ME V+VF+GI N+VFVAV+ CTVVFQIIIVE+LGTFANT+PLSL QW +
Sbjct: 182 FNEISSRDMERVNVFQGILKNYVFVAVLTCTVVFQIIIVEFLGTFANTSPLSLKQWFGSV 361
Query: 997 SVGYVGMPIATYLKQIPV 1014
G +GMPIA LK IPV
Sbjct: 362 LFGVLGMPIAAALKMIPV 415
>TC217853 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase, partial
(10%)
Length = 806
Score = 167 bits (424), Expect = 2e-41
Identities = 80/106 (75%), Positives = 92/106 (86%)
Frame = +2
Query: 909 KWVFFLRGPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTV 968
K +F L GPN+ +VLNTLIFN+FVFCQVFNEINSREME+++VFKGI DN+VFV VI TV
Sbjct: 32 KRIFLLEGPNSDLVLNTLIFNTFVFCQVFNEINSREMEKINVFKGILDNYVFVGVISATV 211
Query: 969 VFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
FQIIIVEYLGTFANTTPL+L QW FCL VG++GMPIA LK+IPV
Sbjct: 212 FFQIIIVEYLGTFANTTPLTLAQWFFCLLVGFLGMPIAARLKKIPV 349
Score = 29.3 bits (64), Expect = 9.1
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +3
Query: 899 VVIWFLQTVGKWVFFLRGP 917
+VIWFLQ+ GK F+LR P
Sbjct: 3 MVIWFLQSRGKESFYLRAP 59
>TC205394 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase, partial
(12%)
Length = 782
Score = 153 bits (387), Expect = 3e-37
Identities = 85/158 (53%), Positives = 97/158 (60%), Gaps = 39/158 (24%)
Frame = +1
Query: 896 YQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFCQV------------------- 936
YQ +VI FLQ+ G +F L GPN+ V NT IFNSFV CQV
Sbjct: 1 YQXMVIXFLQSRGXSIFLLEGPNSXXVXNTXIFNSFVXCQVTFLKS*QFF*EYLLLNS*C 180
Query: 937 --------------------FNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVE 976
FNEINSREME+++VFKGI DN+VFV VI TV FQIIIVE
Sbjct: 181 MYAICIEVSYKMWVDLVLQVFNEINSREMEKINVFKGILDNYVFVGVISATVFFQIIIVE 360
Query: 977 YLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
YLGTFANTTPL+L QW FCL VG++GMPIA LK+IPV
Sbjct: 361 YLGTFANTTPLTLSQWFFCLLVGFMGMPIAARLKKIPV 474
>AW310835 similar to GP|16508162|g type IIB calcium ATPase {Medicago
truncatula}, partial (12%)
Length = 385
Score = 135 bits (340), Expect = 9e-32
Identities = 69/130 (53%), Positives = 89/130 (68%)
Frame = -1
Query: 102 LGSIVEGHDVKKLKFHGGVSGIAEKLSTSTTKGLSGDSEARRIRQEVYGINKFAESEVRS 161
+ S+V GHD K G V GI EKLS S G+ S R Q++YG+N++ E +S
Sbjct: 385 IASVVRGHDYNYYKKIGQVEGIIEKLSASADDGVGQVSIDTR--QDIYGVNRYTEKPSKS 212
Query: 162 FWIFVYEALQDMTLMILAVCAFVSLIVGIATEGWPQGSHDGLGIVASILLVVFVTATSDY 221
F +FV+EA D+TLMIL VCA VS+ +G+ TEGWP+G +DGLGI+ SI LVV VTA SDY
Sbjct: 211 FLMFVWEARHDLTLMILMVCAIVSIAIGLPTEGWPKGVYDGLGIILSIFLVVIVTAISDY 32
Query: 222 RQSLQFKDLD 231
+QS F+DLD
Sbjct: 31 QQSKPFRDLD 2
>TC231696 weakly similar to UP|ACAA_ARATH (Q9SZR1) Potential
calcium-transporting ATPase 10, plasma membrane-type
(Ca(2+)-ATPase isoform 10) , partial (15%)
Length = 824
Score = 134 bits (336), Expect = 3e-31
Identities = 67/152 (44%), Positives = 101/152 (66%), Gaps = 3/152 (1%)
Frame = +1
Query: 866 TDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRG-PN--AGVV 922
TD LM ++P G++ +++IMWRN+L QA+YQ V+ L G + LR PN A V
Sbjct: 1 TDSLMDQSPKGQREPLVSNIMWRNLLIQAMYQVSVLLILNFRGVSLLALRDEPNRPAIKV 180
Query: 923 LNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFA 982
N+LIFN+FV CQVFNE N+R+ ++ ++FKG+ N++F+ ++G TVV QI+I+EYLG F
Sbjct: 181 KNSLIFNAFVLCQVFNEFNARKPDKFNIFKGVTRNYLFMGIVGITVVLQIVIIEYLGKFT 360
Query: 983 NTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
T L+ QW+ + + ++ P+A K IPV
Sbjct: 361 KTAKLNWKQWLISVIIAFISWPLAVVGKLIPV 456
>BE608389 homologue to GP|11066056|g plasma membrane Ca2+-ATPase {Glycine
max}, partial (10%)
Length = 317
Score = 87.0 bits (214), Expect(2) = 2e-29
Identities = 44/63 (69%), Positives = 51/63 (80%)
Frame = +3
Query: 513 NKRGKREILGTPTESAILEFGLSLGGDPQKERQACKLVKVEPFNSQKKRMGVVVELPEGG 572
NK K EILG+PTE+A+LE GLSLGGD KERQ KLVKVEPFNS KKRMGVV++L +GG
Sbjct: 6 NKDEKIEILGSPTETALLELGLSLGGDFLKERQRSKLVKVEPFNSTKKRMGVVLQLADGG 185
Query: 573 LRA 575
+
Sbjct: 186 FHS 194
Score = 61.6 bits (148), Expect(2) = 2e-29
Identities = 28/43 (65%), Positives = 34/43 (78%)
Frame = +1
Query: 572 GLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTID 614
G AHCKGASEI+LAACD V+DS G+VVPLN S N+L + I+
Sbjct: 184 GFIAHCKGASEIILAACDKVVDSSGEVVPLNEYSINHLNNMIE 312
>TC205559 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transporting
ATPase-like protein {Arabidopsis thaliana;} , partial
(14%)
Length = 855
Score = 126 bits (317), Expect = 4e-29
Identities = 64/142 (45%), Positives = 93/142 (65%), Gaps = 3/142 (2%)
Frame = +3
Query: 876 GRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFL---RGPNAGVVLNTLIFNSFV 932
GR+ I +IMWRN+L QA+YQ V+ L G + L R +A V NTLIFN+FV
Sbjct: 3 GRREPLITNIMWRNLLIQAMYQVSVLLVLNFRGISILGLSHDRKAHAIKVKNTLIFNAFV 182
Query: 933 FCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQW 992
CQ+FNE N+R+ +E ++FKG+ N++F+ +IG TVV QI+I+E+LG F +T L+ W
Sbjct: 183 LCQIFNEFNARKPDEFNIFKGVTRNYLFMGIIGLTVVLQIVIIEFLGKFTSTVRLNWKHW 362
Query: 993 IFCLSVGYVGMPIATYLKQIPV 1014
+ + +G +G P+A K IPV
Sbjct: 363 LISVVIGLIGWPLAVIGKLIPV 428
>TC230130 similar to UP|AHM7_ARATH (Q9SH30) Potential copper-transporting
ATPase 3 , partial (43%)
Length = 1409
Score = 97.8 bits (242), Expect = 2e-20
Identities = 59/176 (33%), Positives = 92/176 (51%)
Frame = +1
Query: 650 GVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPDF 709
GV+ + DP++PG +E + + +S I MVTGDN TA +IARE GI
Sbjct: 655 GVLAVSDPLKPGAQEVISILKSMKIKSIMVTGDNFGTASSIAREVGI------------- 795
Query: 710 REKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIG 769
V+A + P K VK L+ + G VA+ GDG ND+PAL AD+G
Sbjct: 796 --------------ENVIAEAKPDQKAEKVKDLQAS-GYTVAMVGDGINDSPALVAADVG 930
Query: 770 LAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL 825
+A+G AGT++A E+AD++++ N ++T R + I+ + L N++ +
Sbjct: 931 MAIG-AGTDIAIEAADIVLMKSNLEDVITAIDLSRKTFSRIRLNYFWALGYNLLGI 1095
Score = 36.2 bits (82), Expect = 0.075
Identities = 33/122 (27%), Positives = 56/122 (45%), Gaps = 11/122 (9%)
Frame = +1
Query: 386 WWWSADDAMEMLEFFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACET 445
W S+ D+ E+ F I+V +V+A P L LA ++ + L++ A E+
Sbjct: 73 WIPSSMDSFELALQFGISV--MVIACPCALGLATPTAVMVGTGVGASQGVLIKGGQALES 246
Query: 446 MGSATTICSDKTGTLTTNHMTVVKTCI-----------CMSSKEVNNKEHGLCSELPDSA 494
I DKTGTLT +V+T + +++ EVN+ EH L + + A
Sbjct: 247 AHKVDCIVFDKTGTLTVGKPVIVRTELLTKMVLQEFYELVAAAEVNS-EHPLAKAVVEYA 423
Query: 495 QK 496
++
Sbjct: 424 KR 429
>BI320368 similar to SP|Q9LU41|ACA Potential calcium-transporting ATPase 9
plasma membrane-type (EC 3.6.3.8) (Ca(2+)-ATPase isoform
9), partial (10%)
Length = 376
Score = 96.7 bits (239), Expect = 5e-20
Identities = 61/127 (48%), Positives = 79/127 (62%), Gaps = 1/127 (0%)
Frame = +3
Query: 443 CETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVNNKEHGLCSELPDSAQKLLLQSI 502
CETMGSATTICSDKTGTLT N MTVV+ + S +VN + S+L A L+ + I
Sbjct: 3 CETMGSATTICSDKTGTLTLNQMTVVEAYV--GSTKVNPPDDS--SKLHPKALSLINEGI 170
Query: 503 FNNTGGEVVVNK-RGKREILGTPTESAILEFGLSLGGDPQKERQACKLVKVEPFNSQKKR 561
NT G V V K G+ E+ G+PTE AIL + + LG + R ++ V PFNS+KKR
Sbjct: 171 AQNTTGNVFVPKDGGETEVSGSPTEKAIL*WAVKLGMNFDVIRSNSTVLHVFPFNSEKKR 350
Query: 562 MGVVVEL 568
GV ++L
Sbjct: 351 GGVALKL 371
>BG789464 similar to GP|16508162|g type IIB calcium ATPase {Medicago
truncatula}, partial (10%)
Length = 395
Score = 90.5 bits (223), Expect = 3e-18
Identities = 51/127 (40%), Positives = 68/127 (53%)
Frame = +1
Query: 4 YLNESFGGVKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLR 63
+LN + K+ + EAL KWR +VKNP+RRFR+ A+L KR A RR Q +R
Sbjct: 4 FLNPEEFELSDKDRSIEALEKWRSAAWLVKNPRRRFRWAADLVKRKHAEDKRRKIQSTIR 183
Query: 64 VAVLVSKAAFQFIQGVQPSDYLVPDDVKAAGFHICADELGSIVEGHDVKKLKFHGGVSGI 123
V QFI + ++Y V + + AGF I D++ S+V GHD K G V GI
Sbjct: 184 TVFNVKWVEGQFISALPQAEYKVSEKTREAGFGIEPDDIASVVRGHDYTNYKKIGQVEGI 363
Query: 124 AEKLSTS 130
EKL S
Sbjct: 364 IEKLRAS 384
>CK606064
Length = 381
Score = 90.1 bits (222), Expect = 4e-18
Identities = 46/110 (41%), Positives = 66/110 (59%)
Frame = +2
Query: 1 MESYLNESFGGVKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQE 60
MES+LN + ++ + E L KWR +VKNP+RRFR+ A+L KR A RR Q
Sbjct: 50 MESFLNPEEFKLSHRDRSIETLEKWRSAAWLVKNPRRRFRWAADLVKRKHAEDKRRXIQS 229
Query: 61 KLRVAVLVSKAAFQFIQGVQPSDYLVPDDVKAAGFHICADELGSIVEGHD 110
+R A+ V +AA QFI + P++Y V + + AGF I D++ S+ GHD
Sbjct: 230 TIRTALTVRRAADQFISVLPPAEYKVSEKTREAGFSIEPDDIASVCRGHD 379
>TC207402 similar to UP|Q941L1 (Q941L1) Copper-transporting P-type ATPase,
partial (19%)
Length = 721
Score = 78.6 bits (192), Expect = 1e-14
Identities = 50/162 (30%), Positives = 80/162 (48%)
Frame = +1
Query: 666 VQVCRSAGIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPDFREKTQEEMFELIPKIQ 725
++ + G+ MVTGDN TA+A+A+E GI
Sbjct: 13 IEGLQKMGVTPVMVTGDNWRTARAVAKEVGIQ---------------------------D 111
Query: 726 VMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESAD 785
V A P K +V+ + G + A+ GDG ND+PAL AD+G+A+G AGT++A E+A+
Sbjct: 112 VRAEVMPAGKADVVRSFQKD-GSIAAMVGDGINDSPALAAADVGMAIG-AGTDIAIEAAE 285
Query: 786 VIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALLV 827
+++ +N ++T R + I+ F + NVVA+ V
Sbjct: 286 YVLMRNNLEDVITAIDLSRKTFSRIRLNYVFAMAYNVVAIPV 411
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,772,668
Number of Sequences: 63676
Number of extensions: 489069
Number of successful extensions: 2621
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 2576
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2607
length of query: 1014
length of database: 12,639,632
effective HSP length: 107
effective length of query: 907
effective length of database: 5,826,300
effective search space: 5284454100
effective search space used: 5284454100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0347b.6


