

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.14
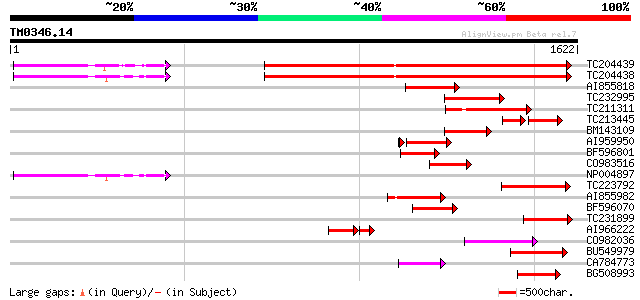
(1622 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 874 0.0
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 870 0.0
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 224 2e-58
TC232995 223 7e-58
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 202 1e-51
TC213445 117 1e-44
BM143109 168 2e-41
AI959950 166 9e-41
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 154 2e-37
CO983516 150 4e-36
NP004897 gag-protease polyprotein 147 3e-35
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 147 3e-35
AI855982 142 2e-33
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 127 4e-29
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 124 3e-28
AI966222 107 3e-28
CO982036 118 2e-26
BU549979 115 2e-25
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 114 5e-25
BG508993 113 6e-25
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 874 bits (2259), Expect = 0.0
Identities = 431/880 (48%), Positives = 598/880 (66%), Gaps = 1/880 (0%)
Frame = +1
Query: 728 CLLSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPF 787
CL S +E + H+R GH +R + ++ VRG+P LK +C CQ GK K+
Sbjct: 2041 CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSH 2220
Query: 788 KAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVF 847
+ +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV F+ K E+ VF
Sbjct: 2221 QKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSETFEVF 2400
Query: 848 STFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKN 907
++Q EK C I R+RSDHG EFEN +F S GI H+FS TPQQNG+VERKN
Sbjct: 2401 KELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKN 2580
Query: 908 RTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFH 967
RTLQE AR ML + + WAE +NTACYI NR+++RR T YE+WK KP++ +FH
Sbjct: 2581 RTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFH 2760
Query: 968 SFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLD 1027
FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 2761 IFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSP 2940
Query: 1028 SDQSKLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIASHPKELIL 1087
+ + + E +V+D K+ E AE + +E+ + R +RI HPKELI+
Sbjct: 2941 ARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDESNINQPDKRSSTRIQKMHPKELII 3120
Query: 1088 GNKDEPVRTRSAFRPSEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNEFSKNDV 1147
G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+EE+ +F +N+V
Sbjct: 3121 GDPNRGVTTRSR----EVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEV 3288
Query: 1148 WSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLE 1207
W LV +P+ +VIGTKW+F+NK NE+G + RNKARLVA+GY+Q EG+D+ ETF PVARLE
Sbjct: 3289 WELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLE 3468
Query: 1208 AIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFKLKKSLYG 1267
+IRLL+ + L+QMDVKSAFLNGY++EEVYV QP GF D +PDHV++LKK+LYG
Sbjct: 3469 SIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKALYG 3648
Query: 1268 LKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGSANQSLCK 1327
LKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVD I+FG + + +
Sbjct: 3649 LKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLR 3828
Query: 1328 EFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAKTPM 1387
F + MQ+EFEMS++GEL YFLG+QV Q + ++ QS+Y K ++KKF M ++ +TP
Sbjct: 3829 HFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKRTPA 4008
Query: 1388 HPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAVK 1447
L K++A V Q LYR MIGSLLYLTASRPD ++V +CAR+Q++P+ +HLT VK
Sbjct: 4009 PTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLTQVK 4188
Query: 1448 RILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKR 1507
RIL+Y+ GT++ G+MY S L GYCD D+AG RKST G C +LG+NL+SW SK+
Sbjct: 4189 RILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWFSKK 4368
Query: 1508 QSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS 1567
Q+ + LSTA+AEYI+A +Q++WMK L++Y + + + +YCDN +AI++SKNP+ HS
Sbjct: 4369 QNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQHS 4548
Query: 1568 RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KAL 1606
R KHI++++H+IRD V V+ LK VDT+ + ADI+ KAL
Sbjct: 4549 RTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKAL 4668
Score = 152 bits (383), Expect = 2e-36
Identities = 127/467 (27%), Positives = 207/467 (44%), Gaps = 20/467 (4%)
Frame = +1
Query: 12 KPPMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERP--VDAEGKKI----PRSEMTA 65
+PP+ DG +EYWK R+ +F D+ W ++ G+E P +D EGK P + T
Sbjct: 34 RPPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTK 213
Query: 66 EQKKLYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLI 125
E+ +L + KA L + + ++ I A +E LK++HEG KVK S+ L
Sbjct: 214 EEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLA 393
Query: 126 QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIE 185
K+E+ M+ E I + + L + T + V +++RSLP+ + VT+IE
Sbjct: 394 TKFENLKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIE 573
Query: 186 LTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESE 245
+D+ M ++ELI L+ EL S+ + + K++A S +E E
Sbjct: 574 EAQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSN----------------DEGE 705
Query: 246 EASEDSDEDE-----LTLISKRLNHIWK--------HKQSKYRGSGKAKGKSESSGQKKS 292
E D D DE + L+ K+ N + H Q+ K + S K S
Sbjct: 706 EDEYDLDTDEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPS 885
Query: 293 SIKEVTCFECKESGHYKSDCPKLKKDKRPKKHFKTKKSLMVTFDESESE-DVDSDGEVQG 351
K + C C+ GH ++CP KKH +K L V ++ESE + DSD +V
Sbjct: 886 HSKGIQCHGCEGYGHIIAECP-----THLKKH---RKGLSVCQSDTESEQESDSDRDVNA 1041
Query: 352 LMTIVKDKEAESKEVPDSDSASEEDPNSDDENEVFASFSTSELKHALSDIMDKYNSLLTK 411
L I + E +SD ++E+ + EL + + K +L +
Sbjct: 1042LTGIFETAE----------------DSSDTDSEI----TFDELAASYRKLCIKSEKILQQ 1161
Query: 412 HKKLKKDFSAASKTPSEHEKIISELKNDNHALVNSNSVLKNQIANLE 458
+LKK + H++ ISELK + L NS L+N +++
Sbjct: 1162EAQLKKVIADLEAEKEAHKEEISELKGEVGFL---NSKLENMTKSIK 1293
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 870 bits (2249), Expect = 0.0
Identities = 429/880 (48%), Positives = 594/880 (66%), Gaps = 1/880 (0%)
Frame = +1
Query: 728 CLLSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPF 787
CL S +E + H+R GH +R + ++ VRG+P LK +C CQ GK K+
Sbjct: 2044 CLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSH 2223
Query: 788 KAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVF 847
+ +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV F+ K ++ VF
Sbjct: 2224 QKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEVF 2403
Query: 848 STFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKN 907
++Q EK C I R+RSDHG EFEN KF S GI H+FS TPQQNG+VERKN
Sbjct: 2404 KELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKN 2583
Query: 908 RTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFH 967
RTLQE AR ML + + WAE +NTACYI NR+++RR T YE+WK KP + +FH
Sbjct: 2584 RTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFH 2763
Query: 968 SFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLD 1027
FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 2764 IFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTP 2943
Query: 1028 SDQSKLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIASHPKELIL 1087
+ + + E +V+D K+ E AE + +E + R RI HPKELI+
Sbjct: 2944 ARKKDVEEDVRTSGDNVADTAKSAENAENSDSATDEPNINQPDKRPSIRIQKMHPKELII 3123
Query: 1088 GNKDEPVRTRSAFRPSEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNEFSKNDV 1147
G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+EE+ +F +N+V
Sbjct: 3124 GDPNRGVTTRSR----EIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEV 3291
Query: 1148 WSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLE 1207
W LV +P+ +VIGTKW+F+NK NE+G + RNKARLVA+GY+Q EG+D+ ETF PVARLE
Sbjct: 3292 WELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLE 3471
Query: 1208 AIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFKLKKSLYG 1267
+IRLL+ + L+QMDVKSAFLNGY++EE YV QP GF D +PDHV++LKK+LYG
Sbjct: 3472 SIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYG 3651
Query: 1268 LKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGSANQSLCK 1327
LKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVD I+FG + + +
Sbjct: 3652 LKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLR 3831
Query: 1328 EFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAKTPM 1387
F + MQ+EFEMS++GEL YFLG+QV Q + ++ QSKY K ++KKF M ++ +TP
Sbjct: 3832 HFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASHKRTPA 4011
Query: 1388 HPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAVK 1447
L K++A V Q LYR MIGSLLYLTASRPD ++V +CAR+Q++P+ +HL VK
Sbjct: 4012 PTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLNQVK 4191
Query: 1448 RILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKR 1507
RIL+Y+ GT++ G+MY S+ L GYCD D+AG RKST G C +LG+NL+SW SK+
Sbjct: 4192 RILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKSTSGGCFYLGTNLISWFSKK 4371
Query: 1508 QSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS 1567
Q+ + LSTA+AEYI+A +Q++WMK L++Y + + + +YCDN +AI++SKNP+ HS
Sbjct: 4372 QNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPVQHS 4551
Query: 1568 RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KAL 1606
R KHI++++H+IRD V V+ L+ VDT+ + ADI+ KAL
Sbjct: 4552 RTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKAL 4671
Score = 153 bits (386), Expect = 7e-37
Identities = 125/468 (26%), Positives = 207/468 (43%), Gaps = 21/468 (4%)
Frame = +1
Query: 12 KPPMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERP--VDAEGKKI----PRSEMTA 65
+PP+ DG +EYWK R+ +F D+ W ++ G+E P +D EGK P + T
Sbjct: 34 RPPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTK 213
Query: 66 EQKKLYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLI 125
E+ +L + KA L + + ++ I A +E LK +HEG KVK S+ L
Sbjct: 214 EEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLA 393
Query: 126 QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIE 185
K+E+ M+ E I + + L + T + V +++RSLP+ + VT+IE
Sbjct: 394 TKFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIE 573
Query: 186 LTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESE 245
+D+ M ++ELI L+ EL S+ + + K++A S +E E
Sbjct: 574 EAQDICNMRVDELIGSLQTFELGLSDRTEKKSKNLAFVSN----------------DEGE 705
Query: 246 EASEDSDEDE-----LTLISKRLNHIWKHKQSKYR--------GSGKAKGKSESSGQKKS 292
E D D DE + L+ K+ N + + + K + S +K S
Sbjct: 706 EDEYDLDTDEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKKSDEKPS 885
Query: 293 SIKEVTCFECKESGHYKSDCPKLKKDKRPKKHFKTKKSLMV-TFDESESE-DVDSDGEVQ 350
K + C C+ GH K++CP K K +K L V D++ESE + DSD +V
Sbjct: 886 HSKGIQCHGCEGYGHIKAECPTHLK--------KQRKGLSVCRSDDTESEQESDSDRDVN 1041
Query: 351 GLMTIVKDKEAESKEVPDSDSASEEDPNSDDENEVFASFSTSELKHALSDIMDKYNSLLT 410
L + + +SD ++E+ + EL + ++ K +L
Sbjct: 1042AL----------------TGRFESAEDSSDTDSEI----TFDELAISYRELCIKSEKILQ 1161
Query: 411 KHKKLKKDFSAASKTPSEHEKIISELKNDNHALVNSNSVLKNQIANLE 458
+ +LKK + HE+ ISELK + L NS L+N +++
Sbjct: 1162QEAQLKKVIANLEAEKEAHEEEISELKGEVGFL---NSKLENMTKSIK 1296
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 224 bits (572), Expect = 2e-58
Identities = 105/153 (68%), Positives = 130/153 (84%)
Frame = -3
Query: 1133 LAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQE 1192
+AM+EE+N+F +N+VW LV+KP++ VIGTKWVFRNKL+E G ++RNKARLVAKGY+Q+E
Sbjct: 461 IAMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEE 282
Query: 1193 GIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDE 1252
GIDY ET+ PVARLE IR+L+++ N L+QMDVKSAFLNG I EEVYV QPPGFE
Sbjct: 281 GIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIP 102
Query: 1253 KNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLE 1285
P HV+KL+K+LYGLKQAPRAWYER+S+FLLE
Sbjct: 101 DKPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>TC232995
Length = 1009
Score = 223 bits (567), Expect = 7e-58
Identities = 110/173 (63%), Positives = 132/173 (75%)
Frame = +2
Query: 1243 VHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTY 1302
V QPPGFE P+HV+KL+K+LYGLKQAPRAWYERLS+FLLE EF RGKVDTTLF K
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 1303 KDDILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYI 1362
+DIL+VQIYVD IIFGS N SLCKEFS MQ+EFEMSMMGELKYFLG+Q+ QT G +I
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 1363 HQSKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLL 1415
+QSKY KEL+K+F M + TPM C L+K+++ + K YR IG ++
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVV 520
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 202 bits (514), Expect = 1e-51
Identities = 116/247 (46%), Positives = 153/247 (60%), Gaps = 2/247 (0%)
Frame = +3
Query: 1248 GFEDEKNPDHVFKLKKSL--YGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDD 1305
GFED++ P HVF + L G+K ++ S ++ K+
Sbjct: 330 GFEDKERPCHVFMV*NKL*ELGMKG*VHF*FQMDSPEE*RTPHYSERLK--------KET 485
Query: 1306 ILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1365
LI+ IYVD IIFG+ ++ +CKEF ++M+ FE SM GELK+ LG+Q+ Q G +IHQ
Sbjct: 486 FLIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIFIHQE 665
Query: 1366 KYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNL 1425
KYTK LK+F M E+ TPMH + I++K++ K Y MI SL YLT+SRPD +
Sbjct: 666 KYTKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTSSRPDIV 845
Query: 1426 FSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT* 1485
F V LCARFQS P+ +H+TAVKRILRYL GTTN L +KK SE+ L GYCDV +AGD+
Sbjct: 846 FVVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYFAGDKVE 1025
Query: 1486 RKSTFGN 1492
RKST N
Sbjct: 1026 RKSTSRN 1046
>TC213445
Length = 705
Score = 117 bits (293), Expect(2) = 1e-44
Identities = 56/98 (57%), Positives = 75/98 (76%)
Frame = +1
Query: 1483 RT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI 1542
+T R+ST C F+GS LVSW SK+Q+++ LSTA+AEYISA Q+ WM+ QL DY +
Sbjct: 400 KTDRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARSYYAQIFWMRQQLFDYGL 579
Query: 1543 LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIR 1580
+IPI CDNT+AI+LSKN IL+SR KHIE+++HF+R
Sbjct: 580 KLDHIPIRCDNTSAINLSKNHILYSRTKHIEIRHHFLR 693
Score = 83.2 bits (204), Expect(2) = 1e-44
Identities = 38/67 (56%), Positives = 49/67 (72%)
Frame = +2
Query: 1410 MIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEY 1469
MI S LYL+ SRP +FSV +C R+Q++P+E+HL+ +KRI+RYL G NLGL Y K S Y
Sbjct: 197 MIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPKNSSY 376
Query: 1470 KLSGYCD 1476
L GY D
Sbjct: 377 NLVGYSD 397
>BM143109
Length = 415
Score = 168 bits (425), Expect = 2e-41
Identities = 84/133 (63%), Positives = 101/133 (75%)
Frame = +1
Query: 1245 QPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD 1304
QPP ++ + P+HVFKLKK LYGLKQA RAWYE LS FLL+ F +GKVDT LF +
Sbjct: 4 QPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLN 183
Query: 1305 DILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1364
DIL+VQIYVD IIFGS N SLCK+FS+ MQ EFEMSMM EL +FLG+Q+ QT G +I Q
Sbjct: 184 DILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFISQ 363
Query: 1365 SKYTKELLKKFNM 1377
SKY K+L+ +F M
Sbjct: 364 SKYCKDLIHRFGM 402
>AI959950
Length = 466
Score = 166 bits (420), Expect(2) = 9e-41
Identities = 84/130 (64%), Positives = 101/130 (77%)
Frame = -1
Query: 1134 AMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEG 1193
AM+EE+++F KN+V LVK P+ V+G KW+F NKL+E G VVR KARLVAKGYSQQEG
Sbjct: 391 AMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEG 212
Query: 1194 IDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEK 1253
IDY +TF VARLE I +L+SF+ N+ L+QMDVKSAFLNG I +EVYV QPPGFE+E
Sbjct: 211 IDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFENET 32
Query: 1254 NPDHVFKLKK 1263
HVFKL K
Sbjct: 31 LHQHVFKLNK 2
Score = 21.2 bits (43), Expect(2) = 9e-41
Identities = 8/16 (50%), Positives = 13/16 (81%)
Frame = -2
Query: 1113 LVSLIEPKSIDEALQD 1128
L+ ++PK IDEA++D
Sbjct: 453 LIFEMKPKHIDEAIKD 406
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 154 bits (390), Expect = 2e-37
Identities = 70/111 (63%), Positives = 94/111 (84%)
Frame = +3
Query: 1118 EPKSIDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVV 1177
EPK+I EA+ D +WI+ M+EE+N+F +N+VW LV+KP++ VIGTKWVFRNKL+E G ++
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 1178 RNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDV 1228
RNKARLVAKGY+Q+EGIDY ET+ PVARLEAIR+L++++ N L+QMDV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>CO983516
Length = 724
Score = 150 bits (379), Expect = 4e-36
Identities = 72/120 (60%), Positives = 92/120 (76%)
Frame = +2
Query: 1200 FVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVF 1259
F PVARLE+IRLL+ + L+QMDVKSAFLNGY++EEVYV QP GF D +PDHV+
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 1260 KLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFG 1319
+LKK+LYGLKQAPRAWYERL+ L + + +G +D TLF K ++++I QIYVD I+FG
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFG 724
>NP004897 gag-protease polyprotein
Length = 1923
Score = 147 bits (372), Expect = 3e-35
Identities = 123/468 (26%), Positives = 205/468 (43%), Gaps = 21/468 (4%)
Frame = +1
Query: 12 KPPMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERP--VDAEGKKI----PRSEMTA 65
+PP+ DG +EYWK R+ +F D+ W ++ +E P +D EGK P + T
Sbjct: 34 RPPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKDWEHPKMLDTEGKPTDGLKPEEDWTK 213
Query: 66 EQKKLYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLI 125
E+ +L + KA L + + ++ I A +E LK +HEG KVK S+ L
Sbjct: 214 EEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLA 393
Query: 126 QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIE 185
K+E+ M+ E I + + L + T + V +++RSLP+ + VT+IE
Sbjct: 394 TKFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIE 573
Query: 186 LTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESE 245
+D+ + ++ELI L+ EL S+ + + K++A S +E E
Sbjct: 574 EAQDICNLRVDELIGSLQTFELGLSDRTEKKSKNLAFVSN----------------DEGE 705
Query: 246 EASEDSDEDE-----LTLISKRLNHIWKHKQSKYR--------GSGKAKGKSESSGQKKS 292
E D D DE + L+ K+ N + + + K + S +K S
Sbjct: 706 EDEYDLDTDEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKRSDEKPS 885
Query: 293 SIKEVTCFECKESGHYKSDCPKLKKDKRPKKHFKTKKSLMV-TFDESESE-DVDSDGEVQ 350
K C C+ GH K++CP K K +K L V D++ESE + DSD +V
Sbjct: 886 HSKGFQCHGCEGYGHIKAECPTHLK--------KQRKGLSVCRSDDTESEQESDSDRDVN 1041
Query: 351 GLMTIVKDKEAESKEVPDSDSASEEDPNSDDENEVFASFSTSELKHALSDIMDKYNSLLT 410
L + + +SD ++E+ + EL + ++ K +L
Sbjct: 1042AL----------------TGRFESAEDSSDTDSEI----TFDELATSYRELCIKSEKILQ 1161
Query: 411 KHKKLKKDFSAASKTPSEHEKIISELKNDNHALVNSNSVLKNQIANLE 458
+ +LKK + HE+ ISELK + L NS L+N +++
Sbjct: 1162QEAQLKKVIANLEAEKEAHEEEISELKGEVGFL---NSKLENMTKSIK 1296
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein, partial
(7%)
Length = 804
Score = 147 bits (372), Expect = 3e-35
Identities = 79/200 (39%), Positives = 126/200 (62%), Gaps = 4/200 (2%)
Frame = +1
Query: 1407 YRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMY--- 1463
+R +IGSL YL SRP+ F+V L +RF PR +H+ A KR+LR +KGT G+++
Sbjct: 22 FRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVLFPFK 201
Query: 1464 KKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISA 1523
K+ + L GY D D+ D KST G V+ +SK+Q I LST +AEY++A
Sbjct: 202 AKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAEYVAA 381
Query: 1524 AICSTQMLWMKHQLEDYQILESN-IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDY 1582
++ + Q +WM + LE+ ++ E + + DN +AI+L+K+P LH R+KHIE+++H+IRD
Sbjct: 382 SLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHYIRDQ 561
Query: 1583 VQKGVLLLKFVDTDHKWADI 1602
V KG + +++ + + AD+
Sbjct: 562 VSKGNVTVEYCKAEEQLADL 621
>AI855982
Length = 484
Score = 142 bits (357), Expect = 2e-33
Identities = 74/165 (44%), Positives = 109/165 (65%)
Frame = +2
Query: 1082 PKELILGNKDEPVRTRSAFRPSEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNE 1141
P + I+G+ + V TR + + L + VS+IEPK+I EA+ D +WI+AM+EE+N+
Sbjct: 2 PLDNIIGDISKGVTTRHSLKD----LCNNMAFVSMIEPKNIKEAIVDDNWIIAMQEELNQ 169
Query: 1142 FSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFV 1201
F +N+VW LV+KP + VI TKWVFRNKL+E ++ +KARLVA+GY+Q +G+DY T+
Sbjct: 170 FERNNVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIIIHKARLVAEGYNQVDGLDYEHTYA 349
Query: 1202 PVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQP 1246
+ARL I + +S+ N L+ SA L+G + EVYV QP
Sbjct: 350 SIARL*VIIMPLSYVYIMNSTLYHYACVSALLHGLLLHEVYVDQP 484
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 127 bits (319), Expect = 4e-29
Identities = 62/130 (47%), Positives = 88/130 (67%)
Frame = -2
Query: 1151 VKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLEAIR 1210
V P +G +WV+ K+ G+V R KARLVAKGY+Q GIDY +TF PVA+L +R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 1211 LLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQ 1270
L ++ + + LHQ+D+K+AFL+G + E++Y+ QPPGF + V KL +SLYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 1271 APRAWYERLS 1280
+PRAW+ + S
Sbjct: 46 SPRAWFGKFS 17
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment), partial
(30%)
Length = 687
Score = 124 bits (312), Expect = 3e-28
Identities = 58/141 (41%), Positives = 92/141 (65%), Gaps = 1/141 (0%)
Frame = +2
Query: 1470 KLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQ 1529
+LSGYCD D+AG R+ST G C F+G NLVSW SK+Q+ + S+A+AEY S A+ + +
Sbjct: 14 QLSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCE 193
Query: 1530 MLWMKHQLEDYQILES-NIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 1588
++W+K L++ + E + +YCDN AA+ ++ NP+ H R KHIE+ HFIR+ + +
Sbjct: 194 LMWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSKEI 373
Query: 1589 LLKFVDTDHKWADIYKALSRG 1609
+ +F+ ++ + DI RG
Sbjct: 374 VTEFIGSNDQPVDILTKSLRG 436
>AI966222
Length = 430
Score = 107 bits (266), Expect(2) = 3e-28
Identities = 49/87 (56%), Positives = 62/87 (70%)
Frame = +1
Query: 912 EMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGC 971
EMART L + KHF AEV+N CY+QN+I +R IL +TPYELWK KPNISYF+ F C
Sbjct: 1 EMARTTLNDNLTPKHF*AEVMNIVCYLQNKIYIRPILKRTPYELWKGRKPNISYFYPFRC 180
Query: 972 VCYVLNTKDRLHKFDAKSSKCLLLGYS 998
C+++NTKD L K D+KS + + YS
Sbjct: 181 KCFIINTKDNLGKIDSKSDCGIFIAYS 261
Score = 38.5 bits (88), Expect(2) = 3e-28
Identities = 20/43 (46%), Positives = 28/43 (64%), Gaps = 1/43 (2%)
Frame = +2
Query: 1001 SKGFRFYNTDAKTIEESIHVRF-DDKLDSDQSKLVEKFADLSI 1042
SK FR YN+ IEE+IH+RF +K + + +L E FADL +
Sbjct: 269 SKAFRVYNSGTLVIEEAIHIRFGKNKPNKELLELDESFADLRL 397
>CO982036
Length = 674
Score = 118 bits (295), Expect = 2e-26
Identities = 74/212 (34%), Positives = 116/212 (53%), Gaps = 5/212 (2%)
Frame = -2
Query: 1302 YKDDILIVQ--IYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEG 1359
YK IL V +YVD II GS+ +L + + + + F + ++G+L YF+ I+V P+
Sbjct: 673 YKTHILTVYLLVYVDIIITGSSC-TLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDL 497
Query: 1360 TYIHQSKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTA 1419
+ ++ + +K I+ +PM TC L K D+ YR ++G+L Y T
Sbjct: 496 LFSLRTSIFEIFCRKPR*QAQPIS-SPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTV 320
Query: 1420 SRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYK---KTSEYKLSGYCD 1476
RP+ F+V+ +F S+P ++H T VKRILRYLKG+ + GL K + + G+CD
Sbjct: 319 IRPEISFAVNKVCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCD 140
Query: 1477 VDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQ 1508
D+A ++ST G FLG NL+SW +Q
Sbjct: 139 ADWASAVDDKRSTSGAAVFLGPNLISWWXXKQ 44
>BU549979
Length = 615
Score = 115 bits (288), Expect = 2e-25
Identities = 58/167 (34%), Positives = 104/167 (61%), Gaps = 3/167 (1%)
Frame = -1
Query: 1433 RFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGN 1492
R+QS+P H K+++RYL+GT + LMYK+T+ ++ GY D D+AG R+ST G
Sbjct: 606 RYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRSTSGY 427
Query: 1493 CQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILES---NIPI 1549
L +VSW S +Q+ I ST + E++ ++ +W+K + ++++S + +
Sbjct: 426 IFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRPLKL 247
Query: 1550 YCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTD 1596
YCDN AA+ ++KN +R+KHI++KY IR+ V++ ++++ V+T+
Sbjct: 246 YCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTE 106
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 114 bits (284), Expect = 5e-25
Identities = 57/134 (42%), Positives = 80/134 (59%)
Frame = +3
Query: 1113 LVSLIEPKSIDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNE 1172
L SL P +I EAL W AM +E+ N W LV P +G +WV+ K+
Sbjct: 3 LSSLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGP 182
Query: 1173 KGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAF 1232
G V R KARLVAKGY+Q GI+Y +TF PV L +RL ++ + + LHQ+D+K+AF
Sbjct: 183 NGKVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAF 362
Query: 1233 LNGYISEEVYVHQP 1246
L+G + E++Y+ QP
Sbjct: 363 LHGDLEEDIYMEQP 404
>BG508993
Length = 374
Score = 113 bits (283), Expect = 6e-25
Identities = 52/123 (42%), Positives = 80/123 (64%), Gaps = 1/123 (0%)
Frame = +1
Query: 1454 KGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIEL 1513
KGT + GL Y ++ YKL G+CD D+AGD RKST G F+G + +W+SK+Q + L
Sbjct: 4 KGTIDFGLFYSPSNNYKLVGFCDSDFAGDVDDRKSTTGFVFFMGDCVFTWSSKKQGIVTL 183
Query: 1514 STAKAEYISAAICSTQMLWMKHQLEDYQILE-SNIPIYCDNTAAISLSKNPILHSRAKHI 1572
T +AEY++A C+ +W++ LE+ Q+L+ + IY DN +A L+KN + H R+KHI
Sbjct: 184 FTCEAEYVAATSCTCHAIWLRRLLEELQLLQKESTKIYVDNRSAQELAKNSVFHERSKHI 363
Query: 1573 EVK 1575
+ +
Sbjct: 364 DTR 372
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,593,989
Number of Sequences: 63676
Number of extensions: 962951
Number of successful extensions: 5867
Number of sequences better than 10.0: 189
Number of HSP's better than 10.0 without gapping: 5521
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5783
length of query: 1622
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1512
effective length of database: 5,635,272
effective search space: 8520531264
effective search space used: 8520531264
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0346.14


