

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
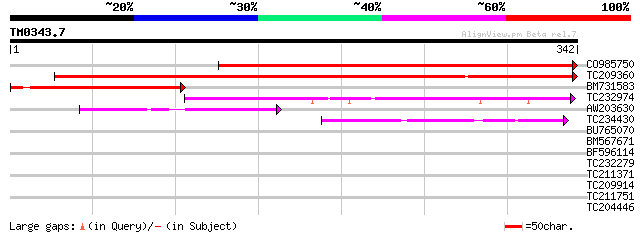
Query= TM0343.7
(342 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO985750 420 e-118
TC209360 UP|DMC1_SOYBN (Q96449) Meiotic recombination protein DM... 342 1e-94
BM731583 similar to SP|Q40134|RA51_ DNA repair protein RAD51 hom... 170 7e-43
TC232974 weakly similar to GB|AAK54457.1|20384750|AY032998 DNA r... 105 4e-23
AW203630 59 2e-09
TC234430 similar to GB|CAC14294.1|11064475|ATH296174 Rad51C prot... 54 1e-07
BU765070 35 0.038
BM567671 33 0.19
BF596114 32 0.32
TC232279 weakly similar to GB|AAR24751.1|38604016|BT010973 At5g0... 29 2.7
TC211371 29 2.7
TC209914 weakly similar to UP|RECA_RICPR (P41079) RecA protein (... 29 3.5
TC211751 weakly similar to UP|Q9KBJ7 (Q9KBJ7) Intracellular alka... 28 4.6
TC204446 similar to UP|ATP4_IPOBA (Q40089) ATP synthase delta' c... 28 6.0
>CO985750
Length = 835
Score = 420 bits (1080), Expect = e-118
Identities = 213/216 (98%), Positives = 214/216 (98%)
Frame = -3
Query: 127 ELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGA 186
ELYGEFRSGKTQLCHT CVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGA
Sbjct: 833 ELYGEFRSGKTQLCHTXCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGA 654
Query: 187 DVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQM 246
DVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQM
Sbjct: 653 DVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQM 474
Query: 247 HLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALR 306
HLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQ+KPIGGNIMAHATTTRLALR
Sbjct: 473 HLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQIKPIGGNIMAHATTTRLALR 294
Query: 307 KGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
KGRGEERICKVISSPCLAEAEARFQI AEGVSDVKD
Sbjct: 293 KGRGEERICKVISSPCLAEAEARFQICAEGVSDVKD 186
>TC209360 UP|DMC1_SOYBN (Q96449) Meiotic recombination protein DMC1 homolog,
complete
Length = 1386
Score = 342 bits (878), Expect = 1e-94
Identities = 177/315 (56%), Positives = 228/315 (72%)
Frame = +2
Query: 28 IEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPM 87
I++L A GI + DVKKL+DAGI T + +K+L IKG+SEAKVDKI EAA KLV
Sbjct: 164 IDKLIAQGINAGDVKKLQDAGIYTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKLVNF 343
Query: 88 GFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTC 147
G+ + S+ ++R+S+I+ITTGS+ LD++L GG+ET +ITE +GEFRSGKTQL HTLCV+
Sbjct: 344 GYITGSDALLKRKSVIRITTGSQALDELLGGGVETSAITEAFGEFRSGKTQLAHTLCVST 523
Query: 148 QLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLL 207
QLP + GG GK YID EGTFRP R++ IA+RFG++ VL+N+ YARAY +HQ LL
Sbjct: 524 QLPTNMRGGNGKVAYIDTEGTFRPDRIVPIAERFGMDPGAVLDNIIYARAYTYEHQYNLL 703
Query: 208 LEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKLADEFGVAIV 267
L A+ M E F ++IVDS AL+R DFSGRGEL+ RQ LA+ L L K+A+EF VA+
Sbjct: 704 LGLAAKMSEEPFRLLIVDSVIALFRVDFSGRGELADRQQKLAQMLSRLIKIAEEFNVAVY 883
Query: 268 ITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVISSPCLAEAE 327
+TNQV+S G VF KP GG+++AHA T RL RKG+GE+RICKV +P L EAE
Sbjct: 884 MTNQVISD-PGGGVFVTDPKKPAGGHVLAHAATVRLMFRKGKGEQRICKVFDAPNLPEAE 1060
Query: 328 ARFQILAEGVSDVKD 342
A FQI A G++D KD
Sbjct: 1061AVFQITAGGIADAKD 1105
>BM731583 similar to SP|Q40134|RA51_ DNA repair protein RAD51 homolog.
[Tomato] {Lycopersicon esculentum}, partial (27%)
Length = 420
Score = 170 bits (431), Expect = 7e-43
Identities = 88/106 (83%), Positives = 96/106 (90%)
Frame = +3
Query: 1 MEQQRHQKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPR 60
MEQQ+ + Q Q+E EEIQHGPLP+EQLQASGIA+ DVKKLKDAGICTVESVAYTPR
Sbjct: 114 MEQQQQRP----QQQDEAEEIQHGPLPVEQLQASGIAATDVKKLKDAGICTVESVAYTPR 281
Query: 61 KDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQIT 106
KDLLQIKGISEAKVDKI+EAASKLVPMGFTSASELH QR++IIQIT
Sbjct: 282 KDLLQIKGISEAKVDKIIEAASKLVPMGFTSASELHAQRDAIIQIT 419
>TC232974 weakly similar to GB|AAK54457.1|20384750|AY032998 DNA repair
protein XRCC3 {Arabidopsis thaliana;} , partial (40%)
Length = 909
Score = 105 bits (261), Expect = 4e-23
Identities = 85/264 (32%), Positives = 125/264 (47%), Gaps = 28/264 (10%)
Frame = +2
Query: 106 TTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDA 165
T G LD+ L GG+ S+TE GE GKTQLC L ++ QLP GG +++I
Sbjct: 44 TLGCPVLDRCLAGGVPCASVTEFVGESGCGKTQLCLQLALSAQLPPSHGGLSASSIFIHT 223
Query: 166 EGTFRPQRLLQIADRF-----GLNGADVLENVAYARAYNTDHQ--------SRLLLEAAS 212
E F +RL ++ F L +D + V + RA ++ H+ LL + S
Sbjct: 224 EFPFPFRRLRHLSRAFRASHPDLPCSDPCDRV-FLRAVHSAHELLNLIPTIETFLLHSKS 400
Query: 213 MMVETRFAVMIVDSATALYRTDFSGRG-ELSARQMHLAKFLRSLQKLADEFGVAIVITNQ 271
R ++++DS AL+R+DF G +L R L++LA FG+A+V+TNQ
Sbjct: 401 PWRPVR--IIVIDSIAALFRSDFENTGSDLRRRSSLFFGISGGLRQLAKRFGIAVVVTNQ 574
Query: 272 VVSQV-DGSAVF---------AGPQVKPIGGNIMAHATTTRLALRKGRGE----ERICKV 317
VV + DG F +G +V P G AH +RL L K E R +V
Sbjct: 575 VVDLIGDGDVSFGSLGNGLYSSGRRVCPALGLAWAHCVNSRLFLSKDEDEPPVKTRKMRV 754
Query: 318 ISSPCLAEAEARFQILAEGVSDVK 341
+ +P L + + I EGV V+
Sbjct: 755 VFAPHLPHSSCEYVIKGEGVFGVE 826
>AW203630
Length = 414
Score = 59.3 bits (142), Expect = 2e-09
Identities = 42/122 (34%), Positives = 62/122 (50%)
Frame = +1
Query: 43 KLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESI 102
KL AG T++++A L++ +SE++ +I+ ASK P + H
Sbjct: 82 KLLAAGYTTLDAIARASPTHLVRDIDVSESEATEILNLASK--PSALERPNGSH------ 237
Query: 103 IQITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMY 162
T +LD IL GGI+ +TE+ G GKTQ+ L V Q+P + GG GKA+Y
Sbjct: 238 ---TAVVGDLDNILGGGIKCKEVTEIGGVPGIGKTQIGIQLAVNVQIPQEYGGLXGKAIY 408
Query: 163 ID 164
ID
Sbjct: 409 ID 414
>TC234430 similar to GB|CAC14294.1|11064475|ATH296174 Rad51C protein
{Arabidopsis thaliana;} , partial (54%)
Length = 789
Score = 53.5 bits (127), Expect = 1e-07
Identities = 44/151 (29%), Positives = 71/151 (46%), Gaps = 2/151 (1%)
Frame = +3
Query: 189 LENVAYARAYNTDHQSRLLLEAASMMVETR-FAVMIVDSATALYRTDFSGRGELSARQMH 247
LEN+ Y + Q L+ E + ++IVDS T +R DF +++ R
Sbjct: 129 LENIFYFXVCSYTEQIALINYLDKFXXENKDVKILIVDSVTFHFRQDFD---DMALRTRL 299
Query: 248 LAKFLRSLQKLADEFGVAIVITNQV-VSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALR 306
L++ L KLA +F +A+V+ NQV ++GS Q+ G+ +H+ T R+ L
Sbjct: 300 LSEMALKLMKLAKKFRLAVVMFNQVPTKHIEGSF-----QLTLALGDSWSHSCTNRIIL- 461
Query: 307 KGRGEERICKVISSPCLAEAEARFQILAEGV 337
G ER + SP L A A + + G+
Sbjct: 462 FWNGNERHAFIAKSPSLKSASAPYSVTTRGI 554
>BU765070
Length = 409
Score = 35.4 bits (80), Expect = 0.038
Identities = 23/57 (40%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Frame = +3
Query: 116 LDGGIETGSITELYGEFRSGKTQLC-HTLCVTCQLPLDQGGGEGKAMYIDAEGTFRP 171
L GG+ G I E+YG SGKT L H + +L G AM +DAE F P
Sbjct: 153 LGGGLPKGRIIEIYGPESSGKTTLALHAIAEVQKL-------GGNAMLVDAEHAFDP 302
>BM567671
Length = 428
Score = 33.1 bits (74), Expect = 0.19
Identities = 22/60 (36%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Frame = +3
Query: 97 VQRESIIQITTGSRELDKILD-GGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGG 155
V +++ ++TGS LD L GG+ G + E+YG SGKT L + Q QGG
Sbjct: 243 VSPKNVPVVSTGSFALDVALGIGGLPKGRVVEIYGPEASGKTTLALHVIAEAQ---KQGG 413
>BF596114
Length = 395
Score = 32.3 bits (72), Expect = 0.32
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = +1
Query: 112 LDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTC 147
+D +L GG+ G +TEL G F K T+C+ C
Sbjct: 127 IDALLRGGLREGQLTELVGSFLVWKDTGMPTVCLNC 234
>TC232279 weakly similar to GB|AAR24751.1|38604016|BT010973 At5g01880
{Arabidopsis thaliana;} , partial (41%)
Length = 703
Score = 29.3 bits (64), Expect = 2.7
Identities = 15/40 (37%), Positives = 22/40 (54%)
Frame = -3
Query: 280 AVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVIS 319
A++ P PI A+ TTTR+ R+ RG R+C +S
Sbjct: 380 ALWKEPYRVPIRELSQANDTTTRIR*RRRRGSSRVCSSVS 261
>TC211371
Length = 677
Score = 29.3 bits (64), Expect = 2.7
Identities = 13/37 (35%), Positives = 23/37 (62%)
Frame = -3
Query: 226 SATALYRTDFSGRGELSARQMHLAKFLRSLQKLADEF 262
SA A Y + SGR L+ ++ +A R+L++ ++EF
Sbjct: 354 SACAKYASTISGRSSLAKSRLSMATHFRTLEEKSEEF 244
>TC209914 weakly similar to UP|RECA_RICPR (P41079) RecA protein (Recombinase
A), partial (39%)
Length = 1105
Score = 28.9 bits (63), Expect = 3.5
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 9/85 (10%)
Frame = +2
Query: 239 GELSARQMHLAKFLRS--LQKLADEFGVA---IVITNQVVSQVDGSAVFAGPQVKPIGGN 293
GE+ M + L S L+KL ++ +V NQV S++ + GP GGN
Sbjct: 8 GEMGDAHMAMQARLMSQALRKLCHSLSLSQCILVFINQVRSKISTFGGYGGPTEVTCGGN 187
Query: 294 IMAHATTTRLALRK----GRGEERI 314
+ + RL +++ +GEE +
Sbjct: 188 ALKFYASVRLNIKRIGFVKKGEETL 262
>TC211751 weakly similar to UP|Q9KBJ7 (Q9KBJ7) Intracellular alkaline serine
protease, partial (27%)
Length = 763
Score = 28.5 bits (62), Expect = 4.6
Identities = 14/38 (36%), Positives = 24/38 (62%), Gaps = 1/38 (2%)
Frame = +2
Query: 111 ELDKILDGGIETGSITELYGEFRSGKTQ-LCHTLCVTC 147
E++ +L G + T SI+ ++GEF + T+ LC +L C
Sbjct: 20 EVNPLLFGLLSTFSISFIFGEFSASSTELLCASLATCC 133
>TC204446 similar to UP|ATP4_IPOBA (Q40089) ATP synthase delta' chain,
mitochondrial precursor , partial (92%)
Length = 954
Score = 28.1 bits (61), Expect = 6.0
Identities = 13/43 (30%), Positives = 24/43 (55%)
Frame = -3
Query: 93 SELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSG 135
S LHV++ + ++ G+ L ++ +GGI G + GE +G
Sbjct: 298 SGLHVRQRRLRRVDVGAHFLPRLREGGIRGGGRRHVGGEAAAG 170
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,844,291
Number of Sequences: 63676
Number of extensions: 116311
Number of successful extensions: 589
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 582
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 585
length of query: 342
length of database: 12,639,632
effective HSP length: 98
effective length of query: 244
effective length of database: 6,399,384
effective search space: 1561449696
effective search space used: 1561449696
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0343.7


