

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.21
(656 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
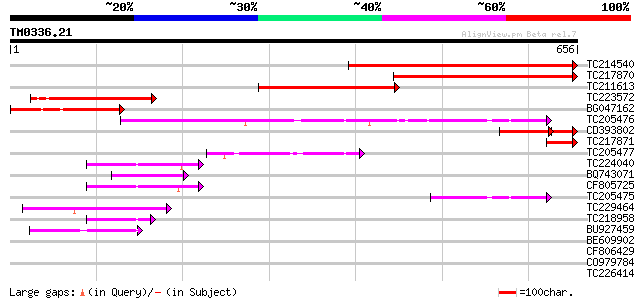
Score E
Sequences producing significant alignments: (bits) Value
TC214540 similar to UP|Q42768 (Q42768) Acetohydroxyacid synthase... 499 e-141
TC217870 homologue to UP|Q42767 (Q42767) Acetohydroxyacid syntha... 397 e-111
TC211613 similar to UP|Q9FUD0 (Q9FUD0) Acetolactate synthase , ... 261 9e-70
TC223572 homologue to UP|Q6T859 (Q6T859) Acetolactate synthase 1... 194 1e-49
BG047162 similar to GP|13958149|gb| acetolactate synthase {Amara... 162 3e-40
TC205476 similar to UP|Q9LF46 (Q9LF46) 2-hydroxyphytanoyl-CoA ly... 143 2e-34
CD393802 similar to GP|1130682|emb| acetohydroxyacid synthase {G... 116 1e-32
TC217871 homologue to UP|Q42767 (Q42767) Acetohydroxyacid syntha... 67 2e-11
TC205477 similar to UP|Q9FNY6 (Q9FNY6) Oxalyl-CoA decarboxylase,... 61 1e-09
TC224040 similar to UP|Q9FVF0 (Q9FVF0) Pyruvate decarboxylase, p... 60 2e-09
BQ743071 similar to PIR|T51575|T51 2-hydroxyphytanoyl-CoA lyase-... 57 2e-08
CF805725 54 2e-07
TC205475 similar to UP|Q9FNY6 (Q9FNY6) Oxalyl-CoA decarboxylase,... 54 2e-07
TC229464 UP|Q7XJ86 (Q7XJ86) Glycinamide ribonucleotide transform... 46 6e-05
TC218958 similar to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 ... 44 3e-04
BU927459 similar to GP|10121330|gb pyruvate decarboxylase {Fraga... 42 7e-04
BE609902 similar to GP|29373077|gb pyruvate decarboxylase 1 {Lot... 42 0.001
CF806429 40 0.003
CO979784 40 0.004
TC226414 homologue to UP|Q84V95 (Q84V95) Pyruvate decarboxylase ... 38 0.013
>TC214540 similar to UP|Q42768 (Q42768) Acetohydroxyacid synthase , partial
(39%)
Length = 1391
Score = 499 bits (1285), Expect = e-141
Identities = 242/264 (91%), Positives = 251/264 (94%)
Frame = +1
Query: 393 PHVSVCADLKLALKGINRILESRGVKGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQY 452
PHVSVCADLKLALKGINR+LESRGV GKLDFR WREELNEQK RFPL +KTFE+ I PQY
Sbjct: 1 PHVSVCADLKLALKGINRVLESRGVAGKLDFRGWREELNEQKRRFPLSYKTFEKEISPQY 180
Query: 453 AIQVLDELTNGNAIISTGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVA 512
AIQVLDELTNG AI+STGVGQHQMWAAQFYKYKRPRQWLTS GLGAMGFGLPAA+GAAVA
Sbjct: 181 AIQVLDELTNGEAIVSTGVGQHQMWAAQFYKYKRPRQWLTSGGLGAMGFGLPAAIGAAVA 360
Query: 513 NPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHT 572
NPG VVVDIDGDGSFMMNVQELATIKVE LPVKILLLNNQHLGMVVQWEDRFYKSNRAHT
Sbjct: 361 NPGAVVVDIDGDGSFMMNVQELATIKVEKLPVKILLLNNQHLGMVVQWEDRFYKSNRAHT 540
Query: 573 YLGDPSNEKEVFPNMLKFADACGIPAARVTKKQDLRAAIQKMLDTPGPYLLDVIVPHQEH 632
YLGDPSNE ++PNMLKFADACGIPAARVTKK+DLRAAIQKML+TPGPYLLDVIVPHQEH
Sbjct: 541 YLGDPSNENAIYPNMLKFADACGIPAARVTKKEDLRAAIQKMLETPGPYLLDVIVPHQEH 720
Query: 633 VLPMIPSNGTFQDVITEGDGRRSY 656
VLPMIPSNGTFQDVITEGDGR SY
Sbjct: 721 VLPMIPSNGTFQDVITEGDGRTSY 792
>TC217870 homologue to UP|Q42767 (Q42767) Acetohydroxyacid synthase ,
partial (32%)
Length = 913
Score = 397 bits (1021), Expect = e-111
Identities = 190/212 (89%), Positives = 205/212 (96%)
Frame = +3
Query: 445 EEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGLP 504
++AI PQ+AI+VLDELTNG+AI+STGVGQHQMWAAQFYKYKRPRQWLTS GLGAMGFGLP
Sbjct: 9 QDAISPQHAIEVLDELTNGDAIVSTGVGQHQMWAAQFYKYKRPRQWLTSGGLGAMGFGLP 188
Query: 505 AAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGMVVQWEDRF 564
AA+GAAVANPG VVVDIDGDGSF+MNVQELATI+VENLPVKILLLNNQHLGMVVQWEDRF
Sbjct: 189 AAIGAAVANPGAVVVDIDGDGSFIMNVQELATIRVENLPVKILLLNNQHLGMVVQWEDRF 368
Query: 565 YKSNRAHTYLGDPSNEKEVFPNMLKFADACGIPAARVTKKQDLRAAIQKMLDTPGPYLLD 624
YKSNRAHTYLGDPS+E E+FPNMLKFADACGIPAARVTKK++LRA IQKMLDTPGPYLLD
Sbjct: 369 YKSNRAHTYLGDPSSESEIFPNMLKFADACGIPAARVTKKEELRAEIQKMLDTPGPYLLD 548
Query: 625 VIVPHQEHVLPMIPSNGTFQDVITEGDGRRSY 656
VIVPHQEHVLPMIPSNG+F+DVITEGDGR Y
Sbjct: 549 VIVPHQEHVLPMIPSNGSFKDVITEGDGRTRY 644
>TC211613 similar to UP|Q9FUD0 (Q9FUD0) Acetolactate synthase , partial
(23%)
Length = 493
Score = 261 bits (666), Expect = 9e-70
Identities = 134/164 (81%), Positives = 142/164 (85%), Gaps = 1/164 (0%)
Frame = +2
Query: 289 VLYAGGGSLNSSEELRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGTVYANYAVD 348
VLY GGGSLNSS+ELRRFVELTG+PVASTLMGLG YP DE SLQMLGMHGTVYANYAVD
Sbjct: 2 VLYVGGGSLNSSDELRRFVELTGIPVASTLMGLGTYPIGDEYSLQMLGMHGTVYANYAVD 181
Query: 349 KSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGI 408
SDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIG NKQ HVSVCADLKLAL GI
Sbjct: 182 NSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGNNKQAHVSVCADLKLALHGI 361
Query: 409 NRILESRGV-KGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQ 451
N ILE + V GKLD +WRE +N QK +FPL +KTF +AI PQ
Sbjct: 362 NMILEKKTVGGGKLDLASWREYINVQKHKFPLGYKTFHDAISPQ 493
>TC223572 homologue to UP|Q6T859 (Q6T859) Acetolactate synthase 1, partial
(15%)
Length = 442
Score = 194 bits (492), Expect = 1e-49
Identities = 103/145 (71%), Positives = 114/145 (78%)
Frame = +3
Query: 25 VLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVATENFTSRFSLDEPRK 84
+ + T+P S+ + P+ +L+I S+S P A A TE F SRF+ EPRK
Sbjct: 18 ITRSTLPL-SHQTLTKPN---HALKIKCSISKPPTAAPFTKEAPTTEPFVSRFASGEPRK 185
Query: 85 GADILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSS 144
GADILVEALERQGVT VFAYPGGASMEIHQALTRSA IRNVLPRHEQGG+FAAEGYARSS
Sbjct: 186 GADILVEALERQGVTTVFAYPGGASMEIHQALTRSAAIRNVLPRHEQGGVFAAEGYARSS 365
Query: 145 GLPGVCIATSGPGATNLVSGLADAL 169
GLPGVCIATSGPGATNLVSGLADAL
Sbjct: 366 GLPGVCIATSGPGATNLVSGLADAL 440
>BG047162 similar to GP|13958149|gb| acetolactate synthase {Amaranthus
retroflexus}, partial (10%)
Length = 414
Score = 162 bits (411), Expect = 3e-40
Identities = 92/134 (68%), Positives = 104/134 (76%), Gaps = 1/134 (0%)
Frame = +2
Query: 1 MAAAPAAKLAFTTVPSPSSRP-QNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTP 59
MAA A K AFT +PS SS Q L+ + FPS PN SS HS+ SL+IS++LS+ T
Sbjct: 23 MAATTAPKPAFTALPSSSSSSSQKPFLRLALQFPSLPN-SSYHSQRPSLKISSALSDAT- 196
Query: 60 TAASASTAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRS 119
A +TA A E+F SRF L+EPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRS
Sbjct: 197 --AKTTTAAAAEDFVSRFGLEEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRS 370
Query: 120 ATIRNVLPRHEQGG 133
A+IRNVLPRHEQGG
Sbjct: 371 ASIRNVLPRHEQGG 412
>TC205476 similar to UP|Q9LF46 (Q9LF46) 2-hydroxyphytanoyl-CoA lyase-like
protein, partial (96%)
Length = 2040
Score = 143 bits (361), Expect = 2e-34
Identities = 131/522 (25%), Positives = 230/522 (43%), Gaps = 23/522 (4%)
Frame = +2
Query: 129 HEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIG 188
+EQ +AA Y +G PGV + SGPG + ++GL++A +++ P + I+G + +G
Sbjct: 197 NEQSAGYAASAYGYLTGRPGVFLTVSGPGCVHGLAGLSNASVNTWPTVMISGSCDQNDVG 376
Query: 189 TDAFQETPIVEVTRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDIQQQL 248
FQE +E T+ TK + I +IP V + A S RPG +D+P D+ Q
Sbjct: 377 RGDFQELNQIEATKPFTKLSVKASHISEIPARVAQVLDWAQSARPGGCYLDLPTDVLHQK 556
Query: 249 AVPNWDQPIKLPGYMARLPRSPS------DALLEQVVRLLLECKKPVLYAGGGSLNSSEE 302
+ + + R +P ++ +EQ V LL ++P++ G G+ + E
Sbjct: 557 ISESEAEKLLSEAENNRSISNPKPEPPLFNSKIEQAVSLLRHAERPLIVFGKGAAYARAE 736
Query: 303 --LRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAFGVRF 360
L + V TG+P T MG G P E + T + A+ K D+ L G R
Sbjct: 737 HVLTKLVNSTGIPFLPTPMGKGILPDNHE-------LAATAARSLAIGKCDVALVVGARL 895
Query: 361 DDRV-TGKLEAFASRAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGINRILES----- 414
+ + G+ ++ K + +D+ EI + ++PH+ + D K ++ +N+ ++
Sbjct: 896 NWLLHFGEPPKWSKDVKFILVDVSEEEI-ELRKPHLGLIGDAKHVIEVLNKEIKDDPFCL 1072
Query: 415 -------RGVKGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAII 467
+ K + E+ +K P F T I + AI L + ++
Sbjct: 1073GSTHPWVEAISNKAKDNVAKMEVQLKKDVVPFNFLTPMRII--RDAIAGLG--SPAPVVV 1240
Query: 468 STGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSF 527
S G + + + PR L + G MG GL + AAVA+P +VV ++GD F
Sbjct: 1241SEGANTMDVGRSVLVQ-TEPRTRLDAGTWGTMGVGLGYCIAAAVASPERLVVAVEGDSGF 1417
Query: 528 MMNVQELATIKVENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLGDPSNEKEVFPNM 587
+ E+ T+ LPV +++ NN V DR + DP+ + PN
Sbjct: 1418GFSAMEVETLVRYQLPVVVIVFNNGG----VYGGDRRRPEEIDGPHKDDPA-PTDFVPNA 1582
Query: 588 LKFA--DACGIPAARVTKKQDLRAAIQKMLDTPGPYLLDVIV 627
A +A G V +L++A+ + P +++V++
Sbjct: 1583GYHALIEAFGGKGYLVGTPDELKSALSESFSARKPAVVNVVI 1708
>CD393802 similar to GP|1130682|emb| acetohydroxyacid synthase {Gossypium
hirsutum}, partial (9%)
Length = 586
Score = 116 bits (290), Expect(2) = 1e-32
Identities = 52/63 (82%), Positives = 60/63 (94%)
Frame = -2
Query: 567 SNRAHTYLGDPSNEKEVFPNMLKFADACGIPAARVTKKQDLRAAIQKMLDTPGPYLLDVI 626
S+RAH YLGDPS+E ++FPNMLKFADACGIPAARVTKK++LRA IQ+MLDTPGPYLLDV+
Sbjct: 585 SHRAHPYLGDPSSESDIFPNMLKFADACGIPAARVTKKEELRAPIQRMLDTPGPYLLDVL 406
Query: 627 VPH 629
VPH
Sbjct: 405 VPH 397
Score = 42.7 bits (99), Expect(2) = 1e-32
Identities = 19/29 (65%), Positives = 22/29 (75%)
Frame = -1
Query: 628 PHQEHVLPMIPSNGTFQDVITEGDGRRSY 656
P + VLPMIP NG+F+DVI EGDGR Y
Sbjct: 400 PLRSTVLPMIPRNGSFKDVIIEGDGRTRY 314
>TC217871 homologue to UP|Q42767 (Q42767) Acetohydroxyacid synthase ,
partial (5%)
Length = 388
Score = 67.4 bits (163), Expect = 2e-11
Identities = 30/35 (85%), Positives = 33/35 (93%)
Frame = +1
Query: 622 LLDVIVPHQEHVLPMIPSNGTFQDVITEGDGRRSY 656
+LDVIVPHQEHVLPMIPSNG+F+DVITEGDGR Y
Sbjct: 1 VLDVIVPHQEHVLPMIPSNGSFKDVITEGDGRTRY 105
>TC205477 similar to UP|Q9FNY6 (Q9FNY6) Oxalyl-CoA decarboxylase, partial
(37%)
Length = 709
Score = 61.2 bits (147), Expect = 1e-09
Identities = 51/196 (26%), Positives = 93/196 (47%), Gaps = 13/196 (6%)
Frame = +3
Query: 228 ATSGRPGPVLIDIPKDIQQQ----------LAVPNWDQPIKLPGYMARLPRSPSDALLEQ 277
A S RPG +D+P D+ Q L + ++ I P ++ R S++ +E+
Sbjct: 30 AQSARPGGCYLDLPTDVLHQKISESEAEKLLTEADKNRSISEP----KIERPLSNSKIEE 197
Query: 278 VVRLLLECKKPVLYAGGGSLNSSEE--LRRFVELTGVPVASTLMGLGAYPTADENSLQML 335
V LL ++P++ G G+ + E L + V TG+P T MG G P D ++L
Sbjct: 198 AVSLLRHAERPLIVFGKGAAYARAEHVLTKLVNSTGIPFLPTPMGKGILP--DTHALA-- 365
Query: 336 GMHGTVYANYAVDKSDLLLAFGVRFDDRV-TGKLEAFASRAKIVHIDIDSAEIGKNKQPH 394
T + A+ K D+ L G R + + G+ ++ K + +D+ EI + ++PH
Sbjct: 366 ---ATAARSLAIGKCDVALVVGARLNWLLHFGEPPKWSKDVKFILVDVSEEEI-ELRKPH 533
Query: 395 VSVCADLKLALKGINR 410
+ + D K ++ +N+
Sbjct: 534 LGLIGDAKHVIEVLNK 581
>TC224040 similar to UP|Q9FVF0 (Q9FVF0) Pyruvate decarboxylase, partial (41%)
Length = 837
Score = 60.5 bits (145), Expect = 2e-09
Identities = 39/144 (27%), Positives = 72/144 (49%), Gaps = 8/144 (5%)
Frame = +3
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
L L + GVTDVF+ PG ++ + L ++N+ +E +AA+GYAR G+ G
Sbjct: 210 LARRLVQVGVTDVFSVPGDFNLTLLDHLIAEPQLKNIGCCNELNAGYAADGYARCRGV-G 386
Query: 149 VCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPI--------VEV 200
C+ T G ++++ +A A +++PLI I G GT+ I +
Sbjct: 387 ACVVTFTVGGLSVINAIAGAYSENLPLICIVGGPNTNDFGTNRILHHTIGLSDFSQELRC 566
Query: 201 TRSITKHNYLVLDIDDIPRIVNEA 224
+++T + +V +I+D +++ A
Sbjct: 567 FQTVTCYQAVVNNIEDAHELIDTA 638
>BQ743071 similar to PIR|T51575|T51 2-hydroxyphytanoyl-CoA lyase-like protein
- Arabidopsis thaliana, partial (18%)
Length = 421
Score = 57.4 bits (137), Expect = 2e-08
Identities = 29/88 (32%), Positives = 49/88 (54%)
Frame = +2
Query: 119 SATIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAI 178
S +R + +EQ +AA Y +G PGV + SGPG + ++GL++A +++ P + I
Sbjct: 155 SLGVRFIAFHNEQSAGYAASAYGYLTGRPGVFLTVSGPGCVHGLAGLSNASVNTWPTVMI 334
Query: 179 TGQVPRRMIGTDAFQETPIVEVTRSITK 206
+G + +G FQE +E T+ TK
Sbjct: 335 SGSCDQNDVGRGDFQELNQIEATKPFTK 418
>CF805725
Length = 571
Score = 54.3 bits (129), Expect = 2e-07
Identities = 38/144 (26%), Positives = 69/144 (47%), Gaps = 8/144 (5%)
Frame = +1
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
L L GV DVF+ PG ++ + L ++ V +E +AA+GYAR+ G+ G
Sbjct: 139 LARRLAETGVRDVFSVPGDFNLTLLDHLIAEPSLNLVGCCNELNAGYAADGYARAKGV-G 315
Query: 149 VCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQE--------TPIVEV 200
C+ T G ++++ +A A +++P+I I G GT+ T +
Sbjct: 316 ACVVTFTVGGLSVLNAIAGAYSENLPVICIVGGPNSNDYGTNRILHHTIGLPDFTQELRC 495
Query: 201 TRSITKHNYLVLDIDDIPRIVNEA 224
++IT +V ++DD +++ A
Sbjct: 496 FQAITCFQAVVNNLDDAHELIDTA 567
>TC205475 similar to UP|Q9FNY6 (Q9FNY6) Oxalyl-CoA decarboxylase, partial
(31%)
Length = 723
Score = 53.9 bits (128), Expect = 2e-07
Identities = 41/143 (28%), Positives = 67/143 (46%), Gaps = 2/143 (1%)
Frame = +1
Query: 487 PRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKI 546
PR L + G MG GL + AAVA PG +VV ++GD F + E+ T+ LPV +
Sbjct: 85 PRTRLDAGTWGTMGVGLGYCIAAAVAEPGRLVVAVEGDSGFGFSAMEVETLVRYQLPVVV 264
Query: 547 LLLNNQHLGMVVQWEDRFYKSNRAHTYLGDPSNEKEVFPNMLKFA--DACGIPAARVTKK 604
++ NN V DR + + DP+ + PN A +A G V
Sbjct: 265 IVFNNGG----VYGGDRRHPEEIDGPHKDDPA-PTDFVPNAGYHALIEAFGGKGYLVGTP 429
Query: 605 QDLRAAIQKMLDTPGPYLLDVIV 627
+L++A+ + P +++V++
Sbjct: 430 DELKSALSESFSARKPAVVNVVI 498
>TC229464 UP|Q7XJ86 (Q7XJ86) Glycinamide ribonucleotide transformylase ,
complete
Length = 1551
Score = 45.8 bits (107), Expect = 6e-05
Identities = 44/182 (24%), Positives = 81/182 (44%), Gaps = 9/182 (4%)
Frame = +2
Query: 15 PSPSSRPQNHVLK--FTIPFPSYPNNSS-PHSRCRSLQISASLSNPTPTAASASTAVATE 71
P PSS P ++K F+ FP P++S P S+ ++L + + +P + + +
Sbjct: 293 PKPSSVPSIPIVKQQFSPKFPRLPSSSLYPSSQSQNLNVPSGAFHPISIVHKEVCSSSCK 472
Query: 72 NF---TSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASME-IHQALTRSATIRNVLP 127
+S S EP++G ++ + R+ VF GG++ IH+A R + +VL
Sbjct: 473 RIWCSSSSSSTAEPKEGHEVRAQVTVRRKKLAVFVSGGGSNFRAIHEASKRGSLHGDVLV 652
Query: 128 RHEQGGIFAAEGYARSSGLPGVC--IATSGPGATNLVSGLADALLDSVPLIAITGQVPRR 185
YAR++G+P + I+ ++LV L +D + L +P
Sbjct: 653 LVTNKSDCGGAEYARNNGIPVILYHISKDESNPSDLVDTLRKFEVDFILLAGYLKLIPVE 832
Query: 186 MI 187
+I
Sbjct: 833 LI 838
>TC218958 similar to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 , partial
(18%)
Length = 711
Score = 43.5 bits (101), Expect = 3e-04
Identities = 26/80 (32%), Positives = 42/80 (52%)
Frame = +1
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
L L GVTDVF+ PG ++ + L + V +E +AA+GYAR+ G+ G
Sbjct: 379 LARRLVEIGVTDVFSVPGDFNLTLLDHLIAEPALHLVGCCNELNAGYAADGYARARGV-G 555
Query: 149 VCIATSGPGATNLVSGLADA 168
C+ T G ++++ +A A
Sbjct: 556 ACVVTFTVGGLSVLNAIAGA 615
>BU927459 similar to GP|10121330|gb pyruvate decarboxylase {Fragaria x
ananassa}, partial (18%)
Length = 421
Score = 42.4 bits (98), Expect = 7e-04
Identities = 33/130 (25%), Positives = 57/130 (43%)
Frame = +3
Query: 24 HVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVATENFTSRFSLDEPR 83
H+ + + F + N++ C+S + ++ AT N +S +L
Sbjct: 30 HIFHYYLVFYYFLMNTNILGDCKSGNSDIGCPPNGTVSVIKNSVPATTNTSSDATLGRH- 206
Query: 84 KGADILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARS 143
L L + GV DVF+ PG ++ + L ++NV +E +AA+GYAR
Sbjct: 207 -----LARRLVQVGVKDVFSVPGDFNLTLLDHLIAEPQLKNVGCCNELNAGYAADGYARC 371
Query: 144 SGLPGVCIAT 153
G+ G C+ T
Sbjct: 372 RGV-GACVVT 398
>BE609902 similar to GP|29373077|gb pyruvate decarboxylase 1 {Lotus
corniculatus}, partial (25%)
Length = 559
Score = 41.6 bits (96), Expect = 0.001
Identities = 26/79 (32%), Positives = 42/79 (52%)
Frame = +2
Query: 497 GAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGM 556
G++G+ + A +G A A P V+ GDGSF + Q+++T+ I L+NN + +
Sbjct: 191 GSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLVNNGGITI 370
Query: 557 VVQWEDRFYKSNRAHTYLG 575
VV+ D Y + TY G
Sbjct: 371 VVEMYDGPYIVDINGTYTG 427
>CF806429
Length = 661
Score = 40.4 bits (93), Expect = 0.003
Identities = 24/79 (30%), Positives = 39/79 (48%)
Frame = -2
Query: 497 GAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGM 556
G++G+ + A +G A A P V+ GDGSF + Q+++T+ I L+NN +
Sbjct: 537 GSIGWSVGATLGYAQAVPEKRVISCIGDGSFQVTAQDVSTMLRNEQKTIIFLINNGGYTI 358
Query: 557 VVQWEDRFYKSNRAHTYLG 575
V+ D Y + Y G
Sbjct: 357 EVEIHDGPYNVIKNWNYTG 301
>CO979784
Length = 725
Score = 39.7 bits (91), Expect = 0.004
Identities = 24/79 (30%), Positives = 39/79 (48%)
Frame = -1
Query: 497 GAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGM 556
G++G+ + A +G A A P V+ GDGSF + Q+++T+ I L+NN +
Sbjct: 545 GSIGWSVGATLGYAQAVPEKRVISCIGDGSFQVTAQDVSTMLRNEQKSIIFLINNGGYTI 366
Query: 557 VVQWEDRFYKSNRAHTYLG 575
V+ D Y + Y G
Sbjct: 365 EVEIHDGPYNVIKNWNYTG 309
>TC226414 homologue to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 , partial
(48%)
Length = 1180
Score = 38.1 bits (87), Expect = 0.013
Identities = 24/79 (30%), Positives = 39/79 (48%)
Frame = +2
Query: 497 GAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGM 556
G++G+ + A +G A A P V+ GDGSF + Q+++T+ I L+NN +
Sbjct: 488 GSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTI 667
Query: 557 VVQWEDRFYKSNRAHTYLG 575
V+ D Y + Y G
Sbjct: 668 EVEIHDGPYNVIKNWNYTG 724
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,894,587
Number of Sequences: 63676
Number of extensions: 326513
Number of successful extensions: 2169
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 2119
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2154
length of query: 656
length of database: 12,639,632
effective HSP length: 103
effective length of query: 553
effective length of database: 6,081,004
effective search space: 3362795212
effective search space used: 3362795212
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0336.21


