

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
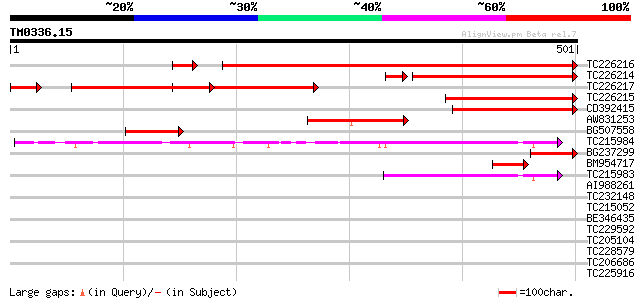
Query= TM0336.15
(501 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226216 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial... 551 e-159
TC226214 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial... 267 2e-77
TC226217 similar to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial (... 207 2e-58
TC226215 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial... 210 1e-54
CD392415 207 7e-54
AW831253 144 7e-35
BG507558 88 8e-18
TC215984 similar to UP|Q9FMP3 (Q9FMP3) Dihydropyrimidinase (Dihy... 85 7e-17
BG237299 79 4e-15
BM954717 similar to GP|18478562|gb| D-hydantoinase {Agrobacteriu... 63 3e-10
TC215983 similar to UP|Q9FMP3 (Q9FMP3) Dihydropyrimidinase (Dihy... 56 4e-08
AI988261 GP|27352107|db hydrolase {Bradyrhizobium japonicum USDA... 33 0.23
TC232148 similar to UP|Q91YE4 (Q91YE4) 67 kDa polymerase-associa... 33 0.30
TC215052 similar to UP|Q9XH69 (Q9XH69) Beta-amylase (Fragment), ... 33 0.39
BE346435 31 1.5
TC229592 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding pr... 31 1.5
TC205104 weakly similar to UP|Q6ZFI7 (Q6ZFI7) Calcium-binding EF... 30 1.9
TC228579 similar to UP|PT13_ARATH (Q8LPL8) Potassium transporter... 30 1.9
TC206686 similar to UP|Q9SCP7 (Q9SCP7) Caffeic acid O-methyltran... 30 3.3
TC225916 weakly similar to GB|AAT41789.1|48310278|BT014806 At3g4... 28 7.3
>TC226216 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial (61%)
Length = 1367
Score = 551 bits (1420), Expect(2) = e-159
Identities = 267/314 (85%), Positives = 291/314 (92%), Gaps = 1/314 (0%)
Frame = +2
Query: 189 KSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLEL-KGNDDPRVY 247
+SFMCPSGI+DFPM TIDHIK GLSVLAKYRRP++VHAEIQQD +N+LEL + N DPR Y
Sbjct: 98 ESFMCPSGIDDFPMATIDHIKAGLSVLAKYRRPIVVHAEIQQDFENHLELNEDNLDPRAY 277
Query: 248 STYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKEAKLRGD 307
TYLNTRPPSWEEAAIK+LV VTKDT GGPLEGAHVHIVHLSDSS SLDLIKEAK GD
Sbjct: 278 LTYLNTRPPSWEEAAIKQLVSVTKDTRKGGPLEGAHVHIVHLSDSSASLDLIKEAKSHGD 457
Query: 308 SISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTV 367
SISVETC HYLAF+SEEIP+GDTRFKCSPPIRDA+NK+KLWEAVLEGHIDLL++DHSPTV
Sbjct: 458 SISVETCPHYLAFTSEEIPNGDTRFKCSPPIRDAYNKDKLWEAVLEGHIDLLSTDHSPTV 637
Query: 368 PELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIASKG 427
PELKL++EGDFLKAWGGISSLQF+LPVTWSYGKK+GLTLEQLSLLWS+KPATLAG+ SKG
Sbjct: 638 PELKLMEEGDFLKAWGGISSLQFNLPVTWSYGKKYGLTLEQLSLLWSKKPATLAGLESKG 817
Query: 428 AIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFKDG 487
I VGNHADIVVW PELEF+L+DDYPVF+KH LSAYMGRRLSGKVL+TFVRGNLVFKDG
Sbjct: 818 TIAVGNHADIVVWQPELEFDLDDDYPVFIKHSELSAYMGRRLSGKVLETFVRGNLVFKDG 997
Query: 488 KHAPAACGVPILAK 501
KHAPA CGV ILAK
Sbjct: 998 KHAPAPCGVQILAK 1039
Score = 28.1 bits (61), Expect(2) = e-159
Identities = 12/22 (54%), Positives = 18/22 (81%)
Frame = +3
Query: 145 LQLKLEAAENELHVDVGFWGGL 166
L+LKL+AAE++++VDVG L
Sbjct: 3 LKLKLDAAEDKIYVDVGMTNNL 68
>TC226214 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial (32%)
Length = 823
Score = 267 bits (683), Expect(2) = 2e-77
Identities = 127/145 (87%), Positives = 138/145 (94%)
Frame = +3
Query: 357 DLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLWSQK 416
DLL+SDHSPTVPELKL++EGDFLKAWGGISSLQF+LPVTWSYGKK+GLTLEQLSLLWS+K
Sbjct: 72 DLLSSDHSPTVPELKLMEEGDFLKAWGGISSLQFNLPVTWSYGKKYGLTLEQLSLLWSKK 251
Query: 417 PATLAGIASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKVLDT 476
PATLAG+ SKGAI VGNHADIVVW PELEF+L+DDYPVF+KH LSAYMGRRLSGKVL+T
Sbjct: 252 PATLAGLESKGAIAVGNHADIVVWQPELEFDLDDDYPVFIKHSELSAYMGRRLSGKVLET 431
Query: 477 FVRGNLVFKDGKHAPAACGVPILAK 501
FVRGNLVFKDGKHAPA CGV ILAK
Sbjct: 432 FVRGNLVFKDGKHAPAPCGVQILAK 506
Score = 40.4 bits (93), Expect(2) = 2e-77
Identities = 16/19 (84%), Positives = 18/19 (94%)
Frame = +1
Query: 333 KCSPPIRDAFNKEKLWEAV 351
KCSPPIRDA+N+EKLW AV
Sbjct: 1 KCSPPIRDAYNREKLWGAV 57
>TC226217 similar to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial (55%)
Length = 970
Score = 207 bits (526), Expect(2) = 2e-58
Identities = 99/127 (77%), Positives = 116/127 (90%)
Frame = +1
Query: 55 SEDLEINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTEWEGFV 114
S +EIN+GKI+S+ EG+ QG KQE VIDYG+AVIMPGLIDVHVHL+EPGRTEWEGF
Sbjct: 307 SGSVEINDGKIISVVEGHAIQGKPKQEEVIDYGDAVIMPGLIDVHVHLNEPGRTEWEGFD 486
Query: 115 TGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNT 174
TGTRAAAAGGVTTVVDMPLN +PTTVS+E L+LKL+AAE++++VDVGFWGGL+PENALNT
Sbjct: 487 TGTRAAAAGGVTTVVDMPLNCYPTTVSKEKLKLKLDAAEDKIYVDVGFWGGLIPENALNT 666
Query: 175 SILEGLL 181
SIL+GLL
Sbjct: 667 SILDGLL 687
Score = 149 bits (376), Expect = 3e-36
Identities = 84/130 (64%), Positives = 97/130 (74%), Gaps = 1/130 (0%)
Frame = +2
Query: 145 LQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCPSGINDFPMTT 204
L L L+ ++ L +D G L + L L G SAGVLG+K FM PSGI+DFPM T
Sbjct: 584 LSLMLQRIKSMLMLDFGEA*SL--KMHLIQVFLMGS*SAGVLGVKPFMWPSGIDDFPMAT 757
Query: 205 IDHIKEGLSVLAKYRRPLLVHAEIQQDSKNNLEL-KGNDDPRVYSTYLNTRPPSWEEAAI 263
IDHIK GLSVLAKYRRP++VHAEIQQD +N+LEL + N DPR Y TYLNTRPPSWEEAAI
Sbjct: 758 IDHIKAGLSVLAKYRRPIVVHAEIQQDFENHLELNEDNLDPRAYLTYLNTRPPSWEEAAI 937
Query: 264 KELVDVTKDT 273
K+LV VTKDT
Sbjct: 938 KQLVSVTKDT 967
Score = 37.7 bits (86), Expect(2) = 2e-58
Identities = 17/28 (60%), Positives = 20/28 (70%)
Frame = +2
Query: 1 MGAAFFPTVITGLPASALSPHKGSFLDQ 28
+ AAFFP +G PA+ L PHKGSFL Q
Sbjct: 233 ISAAFFPIATSGYPANVL*PHKGSFLVQ 316
>TC226215 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial (23%)
Length = 604
Score = 210 bits (535), Expect = 1e-54
Identities = 100/116 (86%), Positives = 108/116 (92%)
Frame = +3
Query: 386 SSLQFDLPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIASKGAIVVGNHADIVVWHPELE 445
SSLQF+LPVTWSYGKK+GLTLEQLSLLWS+KPATLAG+ SKG I VGNHADIVVW PELE
Sbjct: 3 SSLQFNLPVTWSYGKKYGLTLEQLSLLWSKKPATLAGLESKGTIAVGNHADIVVWQPELE 182
Query: 446 FELNDDYPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFKDGKHAPAACGVPILAK 501
F+L+DDYPVF+KH LSAYMGRRLSGKVL+TFVRGNLVFKDGKHAPA CGV ILAK
Sbjct: 183 FDLDDDYPVFIKHSELSAYMGRRLSGKVLETFVRGNLVFKDGKHAPAPCGVQILAK 350
>CD392415
Length = 593
Score = 207 bits (528), Expect = 7e-54
Identities = 97/110 (88%), Positives = 105/110 (95%)
Frame = -1
Query: 392 LPVTWSYGKKHGLTLEQLSLLWSQKPATLAGIASKGAIVVGNHADIVVWHPELEFELNDD 451
LPVTWSYGKKHGLTLEQLSLLWS+KPAT AG+ SKGAI VGNHADIVVW PE+EF+LN+D
Sbjct: 593 LPVTWSYGKKHGLTLEQLSLLWSKKPATFAGLESKGAIAVGNHADIVVWKPEVEFDLNED 414
Query: 452 YPVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFKDGKHAPAACGVPILAK 501
YPVFLKHP+LSAYMGRRLSGKVL+TFVRGNLVFK GKHAP+ACGVPILAK
Sbjct: 413 YPVFLKHPSLSAYMGRRLSGKVLETFVRGNLVFKKGKHAPSACGVPILAK 264
>AW831253
Length = 420
Score = 144 bits (364), Expect = 7e-35
Identities = 77/119 (64%), Positives = 82/119 (68%), Gaps = 30/119 (25%)
Frame = +3
Query: 264 KELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIK----------------------- 300
K+LV VTKDT GGPLEGAHVHIVHLSDSS SLDLIK
Sbjct: 6 KQLVSVTKDTRKGGPLEGAHVHIVHLSDSSASLDLIKVLCALSCLIKIKNGIYQLI*VLM 185
Query: 301 -------EAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVL 352
EAK GDSISVETC HYLAF+SEEIP+GDTRFKCSPPIRDA+NK+KLWEAVL
Sbjct: 186 L*IYLVQEAKSHGDSISVETCPHYLAFTSEEIPNGDTRFKCSPPIRDAYNKDKLWEAVL 362
>BG507558
Length = 434
Score = 88.2 bits (217), Expect = 8e-18
Identities = 41/51 (80%), Positives = 46/51 (89%)
Frame = +3
Query: 103 DEPGRTEWEGFVTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAE 153
+EPGRT WEGF TGTRAAAAGGVTTVVDMPLNNHPTTVS+ETL+LK+ +
Sbjct: 186 NEPGRTVWEGFDTGTRAAAAGGVTTVVDMPLNNHPTTVSKETLKLKVSVID 338
>TC215984 similar to UP|Q9FMP3 (Q9FMP3) Dihydropyrimidinase
(Dihydropyrimidine amidohydrolase) , partial (90%)
Length = 1922
Score = 85.1 bits (209), Expect = 7e-17
Identities = 127/507 (25%), Positives = 217/507 (42%), Gaps = 23/507 (4%)
Frame = +2
Query: 5 FFPTVITGLPASALSPHKGSFLDQGETCEQLLRVGIDFEKRKADPYTLNHSE--DLEINE 62
F T++ LP+S ++ F D G + + L++ G +H + D+ + +
Sbjct: 62 FISTLLLLLPSSLSQSNQ--FCDAGTSSKLLIKGGT--------VVNAHHQQVADVYVED 211
Query: 63 GKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLD-EPGRTEW-EGFVTGTRAA 120
G IV+I + VID +MPG ID H HL+ E TE + F +G AA
Sbjct: 212 GIIVAINPTITV---GDEVTVIDATGKFVMPGGIDPHTHLEFEFMDTETVDDFFSGQAAA 382
Query: 121 AAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELH---VDVGFWGGLVPENALNTSIL 177
AGG T +D + + + +L EA E + +D GF + + + +
Sbjct: 383 LAGGTTMHIDFVIPH------KGSLTAGFEAYEKKAKKSCMDYGFHMAITKWDETVSREM 544
Query: 178 EGLLSA-GVLGLKSFMCPSG---INDFPMTTIDHIKEGLSVLAKYRRPLLVHAE----IQ 229
E ++ G+ K FM G IND + + EG +VHAE +
Sbjct: 545 ELMVKEKGINSFKFFMAYKGALMIND------ELLLEGFKKCKSLGALAMVHAENGDAVY 706
Query: 230 QDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHL 289
+ + +EL G P ++ +RP E A T I +++VH+
Sbjct: 707 EGQEKMIEL-GITGPEGHAL---SRPAVLEGEA-------TARAIRLADFVNTPLYVVHV 853
Query: 290 SDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEI--PDGDT--RFKCSPPIRDAFNKE 345
S +++ I +A+ G + E + LA + PD +T ++ SPPIR + +
Sbjct: 854 M-SIDAMEEIAKARKSGQRVIGEPVASGLALDESWLWHPDFETAAKYVMSPPIRKRGHDK 1030
Query: 346 KLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHG-L 404
L A+ G + L+ +DH K L DF K G++ ++ + + W + G +
Sbjct: 1031ALQAALSTGVLQLVGTDHCAFNSTQKALGIDDFRKIPNGVNGIEERMHLVWDIMVESGQI 1210
Query: 405 TLEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNL-- 461
++ L S + A + I KGAI+ G+ ADI++ +P FE+ H L
Sbjct: 1211SVTDYVRLTSTECARIFNIYPRKGAILPGSDADIIILNPNSTFEIT----AKSHHSKLDT 1378
Query: 462 SAYMGRRLSGKVLDTFVRGNLVFKDGK 488
+ Y GR GK+ T G +V+++ +
Sbjct: 1379NVYEGRSGKGKIEVTIAGGRVVWENNE 1459
>BG237299
Length = 342
Score = 79.3 bits (194), Expect = 4e-15
Identities = 38/41 (92%), Positives = 39/41 (94%)
Frame = +1
Query: 461 LSAYMGRRLSGKVLDTFVRGNLVFKDGKHAPAACGVPILAK 501
LSAYMGRRLSGKVL+TFVRGNLVFKDGKHAPA CGV ILAK
Sbjct: 91 LSAYMGRRLSGKVLETFVRGNLVFKDGKHAPAPCGVQILAK 213
>BM954717 similar to GP|18478562|gb| D-hydantoinase {Agrobacterium sp. IP
I-671}, partial (4%)
Length = 425
Score = 63.2 bits (152), Expect = 3e-10
Identities = 26/32 (81%), Positives = 29/32 (90%)
Frame = +2
Query: 427 GAIVVGNHADIVVWHPELEFELNDDYPVFLKH 458
G I VGNHADIVVW PELEF+L+DDYPVF+KH
Sbjct: 227 GTIAVGNHADIVVWQPELEFDLDDDYPVFIKH 322
>TC215983 similar to UP|Q9FMP3 (Q9FMP3) Dihydropyrimidinase
(Dihydropyrimidine amidohydrolase) , partial (43%)
Length = 938
Score = 55.8 bits (133), Expect = 4e-08
Identities = 44/162 (27%), Positives = 79/162 (48%), Gaps = 4/162 (2%)
Frame = +1
Query: 331 RFKCSPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQF 390
++ SPPIR + + L A+ G + L+ +DH K DF K G++ ++
Sbjct: 97 KYVMSPPIRKRGHDKALQAALSTGVLQLVGTDHCAFNSTQKARGIDDFRKMPNGVNGIEE 276
Query: 391 DLPVTWSYGKKHG-LTLEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPELEFEL 448
+ + W + G +++ + S + A + I KGAI+ G+ ADI++ +P FE+
Sbjct: 277 RMHLVWDIMVESGQISVTDYVRITSTECAKIFNIYPRKGAILPGSDADIIILNPNSSFEM 456
Query: 449 NDDYPVFLKHPNL--SAYMGRRLSGKVLDTFVRGNLVFKDGK 488
+ H L + Y GRR GK+ T V G +V+++ +
Sbjct: 457 S----AKSHHSRLDTNVYEGRRGKGKIEVTIVGGRVVWENNE 570
>AI988261 GP|27352107|db hydrolase {Bradyrhizobium japonicum USDA 110},
partial (20%)
Length = 323
Score = 33.5 bits (75), Expect = 0.23
Identities = 24/92 (26%), Positives = 46/92 (49%), Gaps = 2/92 (2%)
Frame = +1
Query: 14 PASALSPHKGSFLDQGETCEQLLRVGID--FEKRKADPYTLNHSEDLEINEGKIVSITEG 71
P AL P +G + + ++ R+ D F + D T +H + + +G+I +IT
Sbjct: 10 PCRALRPPRGP-RNTPQIIQRRNRMAFDTLFLNARFDGGTRHH---IAVKDGRIAAITSV 177
Query: 72 YGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLD 103
++ ID +A+++PG ++ H+HLD
Sbjct: 178 DQPPAGAE---TIDLKDALVVPGFVEGHIHLD 264
>TC232148 similar to UP|Q91YE4 (Q91YE4) 67 kDa polymerase-associated factor
PAF67, partial (4%)
Length = 470
Score = 33.1 bits (74), Expect = 0.30
Identities = 25/68 (36%), Positives = 35/68 (50%), Gaps = 15/68 (22%)
Frame = -1
Query: 259 EEAAIKELVDVTKDTIHGGP-----------LEGAHVH---IVHLSDSSVSLDLIKEAKL 304
EEA +VDV D +HGGP LEGA VH +VH+ + V++++ E
Sbjct: 236 EEAEDVVVVDVVSDGVHGGPGGVFEETLREGLEGALVHLVDLVHVLLADVAVEVDHEGFY 57
Query: 305 R-GDSISV 311
R GD + V
Sbjct: 56 RVGDEVRV 33
>TC215052 similar to UP|Q9XH69 (Q9XH69) Beta-amylase (Fragment), partial
(92%)
Length = 2020
Score = 32.7 bits (73), Expect = 0.39
Identities = 25/88 (28%), Positives = 39/88 (43%), Gaps = 9/88 (10%)
Frame = +3
Query: 104 EPGRTEWEGFVTGTRAAAAGGVTTVVDMPLN---------NHPTTVSQETLQLKLEAAEN 154
EP R E + GTR+ A G+ V +PL+ NH ++ LKL E
Sbjct: 327 EPVREEKKPSGIGTRSKMANGLRLFVGLPLDAVSYDCKSINHARAIAAGLKALKLLGVEG 506
Query: 155 ELHVDVGFWGGLVPENALNTSILEGLLS 182
V++ W G+V ++A+ G L+
Sbjct: 507 ---VELPIWWGIVEKDAMGQYDWSGYLA 581
>BE346435
Length = 446
Score = 30.8 bits (68), Expect = 1.5
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 2/43 (4%)
Frame = +2
Query: 438 VVWHPELEFELNDDYPVFLKHPNLS--AYMGRRLSGKVLDTFV 478
V W + N D+P+++KH +LS AY G+ LS VL ++
Sbjct: 161 VTWDATVFGVFNPDFPLYIKHEDLS*IAYGGQYLSISVLQLWI 289
>TC229592 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding protein SBP1
(Fragment), partial (91%)
Length = 1267
Score = 30.8 bits (68), Expect = 1.5
Identities = 25/69 (36%), Positives = 35/69 (50%)
Frame = +1
Query: 353 EGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLL 412
+G +DL + PE K LKE DFL+ ISS+ F P + S G GL+L+ L
Sbjct: 256 DGGVDLQLQWNYGLEPERKRLKEQDFLENNSQISSVDFLQPRSVSTGL--GLSLDNTHLT 429
Query: 413 WSQKPATLA 421
+ A L+
Sbjct: 430 STGDSALLS 456
>TC205104 weakly similar to UP|Q6ZFI7 (Q6ZFI7) Calcium-binding EF-hand family
protein-like, partial (26%)
Length = 745
Score = 30.4 bits (67), Expect = 1.9
Identities = 27/98 (27%), Positives = 47/98 (47%), Gaps = 1/98 (1%)
Frame = -3
Query: 205 IDHIKEGLSVLAKY-RRPLLVHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAI 263
++ +++G+ L + RR V AE++ D + LE + R + + + P A+
Sbjct: 410 VELVEDGVVELRELIRRRGDVDAEVRLDEAHGLEGRAELRAREDAVVVEVQHPELLVHAL 231
Query: 264 KELVDVTKDTIHGGPLEGAHVHIVHLSDSSVSLDLIKE 301
+E V + HG ++ H H V LS S SL +KE
Sbjct: 230 REGRLVVHEVAHGAAVDHHHTHSVLLSVSH-SLSEVKE 120
>TC228579 similar to UP|PT13_ARATH (Q8LPL8) Potassium transporter 13
(AtPOT13) (AtKT5), partial (13%)
Length = 974
Score = 30.4 bits (67), Expect = 1.9
Identities = 12/25 (48%), Positives = 17/25 (68%)
Frame = +2
Query: 317 YLAFSSEEIPDGDTRFKCSPPIRDA 341
+L S +E+P GD +FKC+P DA
Sbjct: 356 FLCLSEKELPKGDHKFKCTPFTFDA 430
>TC206686 similar to UP|Q9SCP7 (Q9SCP7) Caffeic acid O-methyltransferase-like
protein (AT3g53140/T4D2_70), partial (89%)
Length = 1298
Score = 29.6 bits (65), Expect = 3.3
Identities = 13/49 (26%), Positives = 24/49 (48%)
Frame = +1
Query: 312 ETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLEGHIDLLT 360
E C H + + +P+G C P + + ++ A+LEG I ++T
Sbjct: 790 EECKHIMQNCHKALPEGGKLIACEPVLPEDSDESHRTRALLEGDIFVMT 936
>TC225916 weakly similar to GB|AAT41789.1|48310278|BT014806 At3g44220
{Arabidopsis thaliana;} , partial (81%)
Length = 986
Score = 28.5 bits (62), Expect = 7.3
Identities = 21/82 (25%), Positives = 33/82 (39%)
Frame = +2
Query: 135 NHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALNTSILEGLLSAGVLGLKSFMCP 194
+HPTT + + L + V + G LVP + L T+ + G L + L K+ P
Sbjct: 527 SHPTTSLASVKTRPTATSSSSLRLTVRYDGKLVPSSLLTTTSM*GALLS*PLAPKATGLP 706
Query: 195 SGINDFPMTTIDHIKEGLSVLA 216
I + + G V A
Sbjct: 707 LAITPLSINWFNAAPSGFDVNA 772
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,339,025
Number of Sequences: 63676
Number of extensions: 309865
Number of successful extensions: 1351
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1338
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1345
length of query: 501
length of database: 12,639,632
effective HSP length: 101
effective length of query: 400
effective length of database: 6,208,356
effective search space: 2483342400
effective search space used: 2483342400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0336.15


