

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
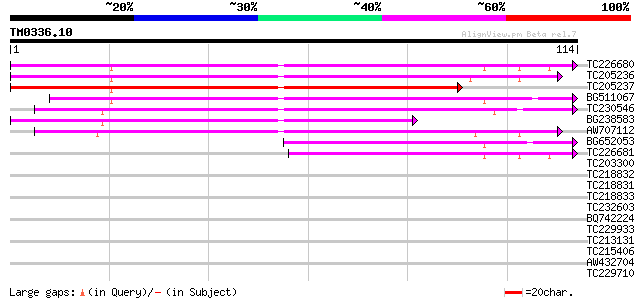
Query= TM0336.10
(114 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226680 weakly similar to UP|SCS2_YEAST (P40075) SCS2 protein, ... 86 3e-18
TC205236 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (82%) 78 5e-16
TC205237 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (58%) 76 3e-15
BG511067 weakly similar to PIR|B86220|B86 protein F22O13.31 [imp... 74 1e-14
TC230546 similar to GB|AAM62506.1|21553413|AY085274 VAMP (vesicl... 64 1e-11
BG238583 similar to GP|8809600|dbj| VAMP (vesicle-associated mem... 54 1e-08
AW707112 53 2e-08
BG652053 similar to GP|20160761|dbj P0504E02.2 {Oryza sativa (ja... 43 2e-05
TC226681 similar to GB|AAQ63968.1|34329673|AY364005 VAP27-1 {Ara... 40 1e-04
TC203300 similar to UP|SPDE_LYCES (Q9ZS45) Spermidine synthase ... 27 1.1
TC218832 similar to UP|Q977S6 (Q977S6) Uncharacterized protein, ... 27 1.4
TC218831 27 1.4
TC218833 27 1.4
TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment)... 26 2.4
BQ742224 26 2.4
TC229933 homologue to UP|SPD2_PEA (Q9ZTR0) Spermidine synthase 2... 26 3.2
TC213131 similar to UP|Q6FJZ0 (Q6FJZ0) Strain CBS138 chromosome ... 26 3.2
TC215406 similar to UP|SPD1_PEA (Q9ZTR1) Spermidine synthase 1 ... 25 4.1
AW432704 25 5.4
TC229710 similar to UP|Q41324 (Q41324) Cationic peroxidase, part... 25 7.1
>TC226680 weakly similar to UP|SCS2_YEAST (P40075) SCS2 protein, partial
(16%)
Length = 1112
Score = 85.5 bits (210), Expect = 3e-18
Identities = 51/123 (41%), Positives = 74/123 (59%), Gaps = 9/123 (7%)
Frame = +3
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M E+L I+P EL F E K+ C L L NKT NYVAF+++ + P+KY V P G++
Sbjct: 126 MSSGELLHIQPQELQFPFELRKQISCSLQLSNKTDNYVAFKVKTTNPKKYCVRPNT-GVV 302
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS----VKDLFTE--NRGKRH-VEEK 111
MP+ T D+IV+ ++ K PP +C D+F +QS+ S KD+ E N+ H VEE
Sbjct: 303 MPRSTCDVIVTMQAQKEAPPDMQCKDKFLLQSVVASPGATTKDITPEMFNKESGHDVEEC 482
Query: 112 ELK 114
+L+
Sbjct: 483 KLR 491
>TC205236 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (82%)
Length = 765
Score = 78.2 bits (191), Expect = 5e-16
Identities = 47/119 (39%), Positives = 69/119 (57%), Gaps = 8/119 (6%)
Frame = +2
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M E+L IEP EL F E K+ C L L NKT +YVAF+++ + P+KY V P GI+
Sbjct: 134 MSTGELLNIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPKKYCVRPNT-GIV 310
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSM----ALSVKDLFTE--NRGKRHVEEK 111
P+ T D+IV+ ++ K P +C D+F +QS+ S KD+ + N+ HV E+
Sbjct: 311 TPRSTCDVIVTMQAQKEAPADMQCKDKFLLQSVKTVDGTSPKDITADMFNKEAGHVVEE 487
>TC205237 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (58%)
Length = 696
Score = 75.9 bits (185), Expect = 3e-15
Identities = 39/93 (41%), Positives = 58/93 (61%), Gaps = 2/93 (2%)
Frame = +3
Query: 1 MDHAEILQIEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M E+L IEP EL F E K+ C L L NKT +YVAF+++ + P+KY V P G++
Sbjct: 165 MSTGELLNIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPKKYCVRPNT-GVV 341
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSM 91
P+ T D+IV+ ++ K P +C D+F +QS+
Sbjct: 342 TPRSTCDVIVTMQAQKEAPADMQCKDKFLLQSV 440
>BG511067 weakly similar to PIR|B86220|B86 protein F22O13.31 [imported] -
Arabidopsis thaliana, partial (27%)
Length = 408
Score = 73.6 bits (179), Expect = 1e-14
Identities = 41/116 (35%), Positives = 68/116 (58%), Gaps = 10/116 (8%)
Frame = +2
Query: 9 IEPAELTFLTE--KKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGTYDI 66
IEPAEL F+ E K+ C + L N T +++AF+++ + P+KY V P GI+ P D
Sbjct: 53 IEPAELRFVFELKKQSSCLVQLANTTDHFIAFKVKTTSPKKYCVRPNI-GIVKPNDKCDF 229
Query: 67 IVSRKSDKLPPPGFECTDRFFIQSMALS--------VKDLFTENRGKRHVEEKELK 114
V+ ++ ++ PP C D+F IQS + D+F+++ GK ++EEK+L+
Sbjct: 230 TVTMQAQRMAPPDMLCKDKFLIQSTVVPFGTTEDDITSDMFSKDSGK-YIEEKKLR 394
>TC230546 similar to GB|AAM62506.1|21553413|AY085274 VAMP (vesicle-associated
membrane protein)-associated protein-like {Arabidopsis
thaliana;} , partial (60%)
Length = 1019
Score = 63.5 bits (153), Expect = 1e-11
Identities = 36/119 (30%), Positives = 67/119 (56%), Gaps = 10/119 (8%)
Frame = +3
Query: 6 ILQIEPAELTFL--TEKKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGT 63
++ + P EL F EK+ +C L ++N T NYVAF+++ + P+KY V P G++ P
Sbjct: 228 LITVNPDELRFQFELEKQTYCDLKVLNNTENYVAFKVKTTSPKKYFVRPNT-GVVHPWDL 404
Query: 64 YDIIVSRKSDKLPPPGFECTDRFFIQSMALSVK--------DLFTENRGKRHVEEKELK 114
I V+ ++ + PP +C D+F +QS ++ D F ++ G++ +E+ +L+
Sbjct: 405 CIIRVTLQAQQEYPPDMQCKDKFLLQSTIVNPNTDVDDLPPDTFNKD-GEKSIEDMKLR 578
>BG238583 similar to GP|8809600|dbj| VAMP (vesicle-associated membrane
protein)-associated protein-like {Arabidopsis thaliana},
partial (35%)
Length = 287
Score = 53.9 bits (128), Expect = 1e-08
Identities = 27/84 (32%), Positives = 49/84 (58%), Gaps = 2/84 (2%)
Frame = +3
Query: 1 MDHAEILQIEPAELTFL--TEKKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGII 58
M ++++ + P EL F EK+ FC L ++N + NYVAF+++ + P+KY V P ++
Sbjct: 39 MTASQLISVSPDELRFHFELEKQTFCDLKVLNNSENYVAFKVKTTSPKKYFVRPNT-AVV 215
Query: 59 MPQGTYDIIVSRKSDKLPPPGFEC 82
P + I V+ ++ + PP +C
Sbjct: 216 QPWDSCIIRVTLQAQREYPPDMQC 287
>AW707112
Length = 433
Score = 53.1 bits (126), Expect = 2e-08
Identities = 35/114 (30%), Positives = 61/114 (52%), Gaps = 8/114 (7%)
Frame = +2
Query: 6 ILQIEPAELTF--LTEKKDFCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGT 63
+L I P EL F +K+ L L NKT Y+ F+++ + P+KY + P II P+ T
Sbjct: 2 LLNI*PLELKFPFKLKKQISYSLQLSNKTNIYITFKVKTTNPKKYYIHPNT-SIITPRST 178
Query: 64 YDIIVSRKSDKLPPPGFECTDRFFIQSMA----LSVKDLFTE--NRGKRHVEEK 111
Y+II + ++ K P + ++F ++S+ K++ T+ N+ H+ EK
Sbjct: 179 YNIIFTIQTQKESPTNIQYENKFLLESIKTIDNTXPKNITTDMFNKRTEHIVEK 340
>BG652053 similar to GP|20160761|dbj P0504E02.2 {Oryza sativa (japonica
cultivar-group)}, partial (12%)
Length = 412
Score = 43.1 bits (100), Expect = 2e-05
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 8/67 (11%)
Frame = +3
Query: 56 GIIMPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS--------VKDLFTENRGKRH 107
GI+ P GT D V+ ++ + PP C D+F +QS + DLF ++ G R
Sbjct: 42 GIVKPHGTCDFTVTMQAQRTAPPDLHCKDKFLVQSAVVPKGTTEDEISSDLFVKDSG-RL 218
Query: 108 VEEKELK 114
V+EK+L+
Sbjct: 219 VDEKKLR 239
>TC226681 similar to GB|AAQ63968.1|34329673|AY364005 VAP27-1 {Arabidopsis
thaliana;} , partial (39%)
Length = 829
Score = 40.4 bits (93), Expect = 1e-04
Identities = 24/65 (36%), Positives = 36/65 (54%), Gaps = 7/65 (10%)
Frame = +3
Query: 57 IIMPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALS----VKDLFTE--NRGKRH-VE 109
++MP+ T D+IV K PP +C D+F +QS+ S KD+ E N+ H VE
Sbjct: 3 VVMPRSTCDVIVXCXXQKEAPPDMQCKDKFLLQSVVASPGATTKDITPEMFNKESGHDVE 182
Query: 110 EKELK 114
E +L+
Sbjct: 183 ECKLR 197
>TC203300 similar to UP|SPDE_LYCES (Q9ZS45) Spermidine synthase (Putrescine
aminopropyltransferase) (SPDSY) , partial (68%)
Length = 1308
Score = 27.3 bits (59), Expect = 1.1
Identities = 14/41 (34%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Frame = +1
Query: 59 MPQGTYDIIVSRKSDKLPPPGFEC---TDRFFIQSMALSVK 96
+P+GTYD+I+ SD P G D F++S+A +++
Sbjct: 643 VPEGTYDVII---SDAFPAMGHSADVLADECFLESVAKALR 756
>TC218832 similar to UP|Q977S6 (Q977S6) Uncharacterized protein, partial
(13%)
Length = 687
Score = 26.9 bits (58), Expect = 1.4
Identities = 20/70 (28%), Positives = 28/70 (39%), Gaps = 3/70 (4%)
Frame = +2
Query: 20 KKDFCKLHL-INKTTNYVAFRLRLSKPEKY--GVTPREGGIIMPQGTYDIIVSRKSDKLP 76
K F L L ++ N +FR +S Y G+TP G ++ P R S +P
Sbjct: 59 KLSFSSLRLDSDRHRNGFSFRFSVSLLNVYS*GITPTGGAVVNPSPPPSATTRRPSRPVP 238
Query: 77 PPGFECTDRF 86
P C F
Sbjct: 239 PLFHACFQEF 268
>TC218831
Length = 559
Score = 26.9 bits (58), Expect = 1.4
Identities = 20/70 (28%), Positives = 28/70 (39%), Gaps = 3/70 (4%)
Frame = +1
Query: 20 KKDFCKLHL-INKTTNYVAFRLRLSKPEKY--GVTPREGGIIMPQGTYDIIVSRKSDKLP 76
K F L L ++ N +FR +S Y G+TP G ++ P R S +P
Sbjct: 52 KLSFSSLRLDSDRHRNGFSFRFSVSLLNVYS*GITPTGGAVVNPSPPPSATTRRPSRPVP 231
Query: 77 PPGFECTDRF 86
P C F
Sbjct: 232 PLFHACFQEF 261
>TC218833
Length = 577
Score = 26.9 bits (58), Expect = 1.4
Identities = 20/70 (28%), Positives = 28/70 (39%), Gaps = 3/70 (4%)
Frame = +3
Query: 20 KKDFCKLHL-INKTTNYVAFRLRLSKPEKY--GVTPREGGIIMPQGTYDIIVSRKSDKLP 76
K F L L ++ N +FR +S Y G+TP G ++ P R S +P
Sbjct: 21 KLSFSSLRLDSDRHRNGFSFRFSVSLLNVYS*GITPTGGAVVNPSPPPSATTRRPSRPVP 200
Query: 77 PPGFECTDRF 86
P C F
Sbjct: 201PLFHACFQEF 230
>TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment), partial
(4%)
Length = 1123
Score = 26.2 bits (56), Expect = 2.4
Identities = 18/62 (29%), Positives = 29/62 (46%), Gaps = 4/62 (6%)
Frame = -3
Query: 23 FCKLHLINKTTNYVAFRLRLSKPEKYGVTPREGGIIMPQGTYDIIVSRKSDKL----PPP 78
FC L TNY+ ++LRL P+ Y ++ G + P + + + KS + PP
Sbjct: 578 FCHL-----CTNYIFYQLRLGSPQNYIISILLGFVSTPFLSSSLQTALKSPTMHHLSPPI 414
Query: 79 GF 80
F
Sbjct: 413 SF 408
>BQ742224
Length = 420
Score = 26.2 bits (56), Expect = 2.4
Identities = 20/70 (28%), Positives = 28/70 (39%), Gaps = 3/70 (4%)
Frame = +3
Query: 20 KKDFCKLHL-INKTTNYVAFRLRLSKPEKY--GVTPREGGIIMPQGTYDIIVSRKSDKLP 76
K F L L ++ N +FR +S Y G+TP G ++ P R S +P
Sbjct: 21 KLSFSSLRLDSDRHRNGFSFRFSVSLLNVYS*GITPTGGAVVNPSPPPSATTRRPSLPVP 200
Query: 77 PPGFECTDRF 86
P C F
Sbjct: 201PLFHACFQEF 230
>TC229933 homologue to UP|SPD2_PEA (Q9ZTR0) Spermidine synthase 2
(Putrescine aminopropyltransferase 2) (SPDSY 2) ,
partial (42%)
Length = 814
Score = 25.8 bits (55), Expect = 3.2
Identities = 12/38 (31%), Positives = 23/38 (59%)
Frame = +2
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALSVK 96
+P+GTYD ++ SD + P E ++ F S+A +++
Sbjct: 5 VPEGTYDAVIVDSSDPI-GPAQELFEKPFFSSVAKALR 115
>TC213131 similar to UP|Q6FJZ0 (Q6FJZ0) Strain CBS138 chromosome M complete
sequence, partial (97%)
Length = 549
Score = 25.8 bits (55), Expect = 3.2
Identities = 13/58 (22%), Positives = 27/58 (46%), Gaps = 3/58 (5%)
Frame = +3
Query: 55 GGIIMPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSM---ALSVKDLFTENRGKRHVE 109
G + P G ++ ++ + LPP F + R + A+ ++ L T G++ +E
Sbjct: 294 GKVTRPHGNSGVVRAKFKNNLPPKSFGASVRVMLYPSSI*AVPIRSLATGTNGRKAME 467
>TC215406 similar to UP|SPD1_PEA (Q9ZTR1) Spermidine synthase 1 (Putrescine
aminopropyltransferase 1) (SPDSY 1) , partial (91%)
Length = 1362
Score = 25.4 bits (54), Expect = 4.1
Identities = 12/38 (31%), Positives = 23/38 (59%)
Frame = +3
Query: 59 MPQGTYDIIVSRKSDKLPPPGFECTDRFFIQSMALSVK 96
+P+GTYD ++ SD + P E ++ F S+A +++
Sbjct: 609 VPEGTYDAVIVDSSDPI-GPAQELFEKPFFASVAKALR 719
>AW432704
Length = 432
Score = 25.0 bits (53), Expect = 5.4
Identities = 8/21 (38%), Positives = 17/21 (80%)
Frame = -3
Query: 23 FCKLHLINKTTNYVAFRLRLS 43
FC ++ +++T+N++AF + LS
Sbjct: 73 FCAVNKLSQTSNHIAFNVSLS 11
>TC229710 similar to UP|Q41324 (Q41324) Cationic peroxidase, partial (62%)
Length = 742
Score = 24.6 bits (52), Expect = 7.1
Identities = 13/35 (37%), Positives = 21/35 (59%), Gaps = 3/35 (8%)
Frame = -3
Query: 64 YDIIVSRKSDKLPPP---GFECTDRFFIQSMALSV 95
+ ++VSR ++ LPPP F C D +SM +S+
Sbjct: 365 FGLVVSRGANVLPPPVLGQFPCNDFAKEESMLVSL 261
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,503,331
Number of Sequences: 63676
Number of extensions: 50834
Number of successful extensions: 254
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 248
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 248
length of query: 114
length of database: 12,639,632
effective HSP length: 90
effective length of query: 24
effective length of database: 6,908,792
effective search space: 165811008
effective search space used: 165811008
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0336.10


