

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0335a.3
(479 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
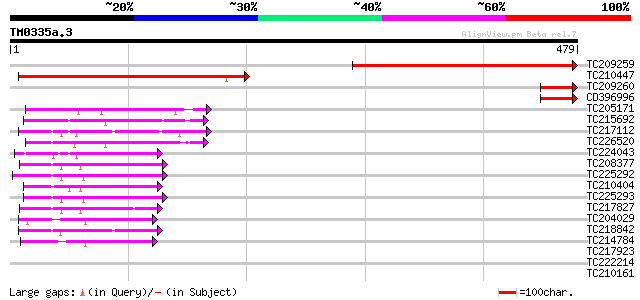
Score E
Sequences producing significant alignments: (bits) Value
TC209259 342 2e-94
TC210447 weakly similar to UP|Q8WUS8 (Q8WUS8) NAD(P) dependent s... 331 4e-91
TC209260 homologue to UP|Q9XHP4 (Q9XHP4) Peroxisomal copper-cont... 60 3e-09
CD396996 54 2e-07
TC205171 similar to PIR|E86431|E86431 T5I8.7 protein - Arabidops... 52 8e-07
TC215692 similar to UP|GAE2_CYATE (O65781) UDP-glucose 4-epimera... 51 1e-06
TC217112 weakly similar to UP|P93188 (P93188) NADPH-dependent HC... 50 2e-06
TC226520 homologue to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimera... 50 3e-06
TC224043 similar to UP|Q6VY19 (Q6VY19) NADPH HC toxin reductase ... 46 3e-05
TC208377 similar to GB|AAM64538.1|21592589|AY086975 cinnamoyl-Co... 45 7e-05
TC225292 weakly similar to GB|AAM64538.1|21592589|AY086975 cinna... 44 2e-04
TC210404 similar to UP|Q8H1H5 (Q8H1H5) Cinnamoyl CoA reductase 2... 44 2e-04
TC225293 weakly similar to GB|AAM64538.1|21592589|AY086975 cinna... 44 2e-04
TC217827 43 3e-04
TC204029 homologue to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydr... 43 3e-04
TC218842 similar to UP|Q9SPJ5 (Q9SPJ5) Dihydroflavonol-4-reducta... 42 6e-04
TC214784 similar to UP|Q9LZI2 (Q9LZI2) DTDP-glucose 4-6-dehydrat... 42 8e-04
TC217923 UP|Q9SPJ5 (Q9SPJ5) Dihydroflavonol-4-reductase DFR1, co... 41 0.001
TC222214 weakly similar to UP|Q89HR7 (Q89HR7) Bll5923 protein , ... 40 0.002
TC210161 UP|Q9SDZ2 (Q9SDZ2) 2'-hydroxy isoflavone/dihydroflavono... 40 0.002
>TC209259
Length = 706
Score = 342 bits (877), Expect = 2e-94
Identities = 161/190 (84%), Positives = 171/190 (89%)
Frame = +1
Query: 290 LPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKE 349
LPKTSL V+ LVLGRIC VYTILYPWL+RWWLPQPFILPSEVHKVGVTHYFSYLKAKE
Sbjct: 1 LPKTSLPVERXLVLGRICWAVYTILYPWLNRWWLPQPFILPSEVHKVGVTHYFSYLKAKE 180
Query: 350 EIGYVPMVSSREGMASTISYWQQRKMITLDGPTIYTWLFCVIGMISLFCAAFLPDVGIVF 409
EIGY PMV+SREGMA TISYWQ+RK TLDGPTIY WLFCVIGMISLFC AFLPD+GI+
Sbjct: 181 EIGYAPMVTSREGMALTISYWQERKRTTLDGPTIYAWLFCVIGMISLFCGAFLPDIGIMS 360
Query: 410 LLRATSLFVFRSMWMIRLVFILATAAHVFEAIYAWYLAKRVDHANARGWFWQTFALGYFS 469
LLR T LFVFRSMW+ RLVF+LATAAH+ EAIYAWYLAKRVD ANARGWFWQTFALG+FS
Sbjct: 361 LLRTTCLFVFRSMWVTRLVFLLATAAHIAEAIYAWYLAKRVDPANARGWFWQTFALGFFS 540
Query: 470 LRFLLKRARK 479
LR LLKR RK
Sbjct: 541 LRLLLKRPRK 570
>TC210447 weakly similar to UP|Q8WUS8 (Q8WUS8) NAD(P) dependent steroid
dehydrogenase-like, partial (18%)
Length = 595
Score = 331 bits (849), Expect = 4e-91
Identities = 163/197 (82%), Positives = 175/197 (88%), Gaps = 2/197 (1%)
Frame = +2
Query: 8 GIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIV 67
GIEGKT VVTGGLGFVGS LCLEL+RRGA EVRAFDLR SSPW LKD GV CIQGD+
Sbjct: 5 GIEGKTFVVTGGLGFVGSGLCLELIRRGAVEVRAFDLRLSSPWSRPLKDKGVLCIQGDVA 184
Query: 68 RKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYC 127
RKEDVE ALRGADCVFHLAAFGMSGKEMLQ RVD+VNI GTCHV+DACL+LGIKRLVYC
Sbjct: 185 RKEDVERALRGADCVFHLAAFGMSGKEMLQFGRVDEVNINGTCHVIDACLYLGIKRLVYC 364
Query: 128 STYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDA--GNCL 185
ST NVVF GQ+I+NGNE+LPYFPIDHHV+PYGRSKSIAEQLVLKNNAR K+D+ + L
Sbjct: 365 STCNVVFGGQQIINGNETLPYFPIDHHVNPYGRSKSIAEQLVLKNNARTLKSDSSGNHRL 544
Query: 186 YTCAVRPAAIYGPGEDR 202
YTCAVRPAAIYGPGEDR
Sbjct: 545 YTCAVRPAAIYGPGEDR 595
>TC209260 homologue to UP|Q9XHP4 (Q9XHP4) Peroxisomal copper-containing amine
oxidase, partial (11%)
Length = 521
Score = 59.7 bits (143), Expect = 3e-09
Identities = 27/31 (87%), Positives = 28/31 (90%)
Frame = +1
Query: 449 RVDHANARGWFWQTFALGYFSLRFLLKRARK 479
R+D ANARGWFWQTFALG FSLR LLKRARK
Sbjct: 232 RMDPANARGWFWQTFALGMFSLRLLLKRARK 324
>CD396996
Length = 450
Score = 53.9 bits (128), Expect = 2e-07
Identities = 25/31 (80%), Positives = 26/31 (83%)
Frame = -1
Query: 449 RVDHANARGWFWQTFALGYFSLRFLLKRARK 479
RVD A AR WFW TFALG+FSLR LLKRARK
Sbjct: 177 RVDPAYARRWFWPTFALGFFSLRLLLKRARK 85
>TC205171 similar to PIR|E86431|E86431 T5I8.7 protein - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (97%)
Length = 1690
Score = 51.6 bits (122), Expect = 8e-07
Identities = 45/167 (26%), Positives = 74/167 (43%), Gaps = 10/167 (5%)
Frame = +2
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDL-RDSSPWFAILKD-----NGVRCIQGDIV 67
++VTGG G++GS L L+R +L R + +L+D ++ I D+
Sbjct: 452 VLVTGGAGYIGSHATLRLLRENYRVTIVDNLSRGNLGAVRVLQDLFPEPGRLQFIYADLG 631
Query: 68 RKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLV 125
KE V D V H AA G+ L + + T VL++ G+K L+
Sbjct: 632 DKESVNKIFSENKFDAVMHFAAVAYVGESTLDPLKYYHNITSNTLLVLESMAKYGVKTLI 811
Query: 126 YCSTYNVVFAGQR--IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL 170
Y ST ++ I+ E P ++PYG++K +AE ++L
Sbjct: 812 YSSTCATYGEPEKMPIIETTEQKP-------INPYGKAKKMAEDIIL 931
>TC215692 similar to UP|GAE2_CYATE (O65781) UDP-glucose 4-epimerase GEPI48
(Galactowaldenase) (UDP-galactose 4-epimerase) , partial
(97%)
Length = 1518
Score = 50.8 bits (120), Expect = 1e-06
Identities = 48/167 (28%), Positives = 77/167 (45%), Gaps = 10/167 (5%)
Frame = +2
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
KT++VTGG G++GS L+L+ G V +L +SS AI + + G+ +
Sbjct: 188 KTVLVTGGAGYIGSHTVLQLLLGGFRAVVLDNLENSSE-VAIHRVRELAGEFGNNLSFHK 364
Query: 72 VESALRGA----------DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGI 121
V+ R A D V H A G+ + + N+TGT +L+ G
Sbjct: 365 VDLRDRAALDQIFSSTQFDAVIHFAGLKAVGESVQKPLLYYNNNLTGTITLLEVMAAHGC 544
Query: 122 KRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
K+LV+ S+ V+ + V E P ++PYGR+K I E++
Sbjct: 545 KKLVFSSS-ATVYGWPKEVPCTEEFPL----SAMNPYGRTKLIIEEI 670
>TC217112 weakly similar to UP|P93188 (P93188) NADPH-dependent HC-toxin
reductase , partial (33%)
Length = 1054
Score = 50.4 bits (119), Expect = 2e-06
Identities = 55/187 (29%), Positives = 88/187 (46%), Gaps = 24/187 (12%)
Frame = +1
Query: 8 GIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILK-----DNGVR 60
G EG + VTGG G++GS L +L+ +G + V A DL++ S +LK + +
Sbjct: 43 GEEGCKVCVTGGSGYIGSWLIKKLLAKG-YTVHATLRDLKNESK-VGLLKSLPQSEGKLV 216
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG 120
+ DI D + A+ G VFH+A M + Q + + GT + +C+ G
Sbjct: 217 LFEADIYNPNDFDLAIEGCKFVFHVAT-PMIHEPGSQYKDTSEAAVAGTKSIFLSCVRAG 393
Query: 121 -IKRLVYCSTYNVVFAGQRIVNG---------------NESLPY-FPIDHHVDPYGRSKS 163
+KRL+Y T +VV A +G N+SL Y + D + Y SK+
Sbjct: 394 TVKRLIY--TASVVSASPLKEDGSGFKDAMDENCWTPLNDSLAYIYRDDPFLKGYTYSKT 567
Query: 164 IAEQLVL 170
++E+ VL
Sbjct: 568 LSERHVL 588
>TC226520 homologue to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase
(Galactowaldenase) (UDP-galactose 4-epimerase) ,
complete
Length = 1290
Score = 49.7 bits (117), Expect = 3e-06
Identities = 40/166 (24%), Positives = 76/166 (45%), Gaps = 11/166 (6%)
Frame = +2
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAI---------LKDNGVRCIQG 64
++VTGG GF+G+ ++L++ G V D D+S A+ L ++ QG
Sbjct: 56 ILVTGGAGFIGTHTVVQLLKAG-FSVSIIDNFDNSVMEAVDRVRQVVGPLLSQNLQFTQG 232
Query: 65 DIVRKEDVESALRGA--DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIK 122
D+ ++D+E D V H A + + + R N+ GT ++ + K
Sbjct: 233 DLRNRDDLEKLFSKTTFDAVIHFAGLKAVAESVAKPRRYFDFNLVGTINLYEFMAKYNCK 412
Query: 123 RLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
++V+ S+ V ++I + F + ++PYGR+K E++
Sbjct: 413 KMVFSSSATVYGQPEKIPCEED----FKL-QAMNPYGRTKLFLEEI 535
>TC224043 similar to UP|Q6VY19 (Q6VY19) NADPH HC toxin reductase (Fragment),
partial (32%)
Length = 575
Score = 46.2 bits (108), Expect = 3e-05
Identities = 36/134 (26%), Positives = 64/134 (46%), Gaps = 9/134 (6%)
Frame = +1
Query: 5 ENEGIEGKTLVVTGGLGFVGSALCLELVRRG--AHEVRAFDLRDSSPWFAILK------D 56
E EGI K + VTGG ++GS L +L+++G H + +D S +L+ D
Sbjct: 1 EREGIRCK-VCVTGGASYIGSCLVKKLLQKGYTVHSTLR-NFKDESK-IGLLRGLPHAND 171
Query: 57 NGVRCIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDAC 116
+ + DI + ++ E A++G + VFH+A + L + I G + C
Sbjct: 172 ERLVLFEADIYKPDEYEPAIQGCEIVFHVATPYEHQSDSLLFKNTSEAAIAGVKSIAKYC 351
Query: 117 LHLGI-KRLVYCST 129
+ G +RL+Y ++
Sbjct: 352 IKSGTERRLIYTAS 393
>TC208377 similar to GB|AAM64538.1|21592589|AY086975 cinnamoyl-CoA
reductase-like protein {Arabidopsis thaliana;} , partial
(98%)
Length = 1087
Score = 45.1 bits (105), Expect = 7e-05
Identities = 41/131 (31%), Positives = 66/131 (50%), Gaps = 6/131 (4%)
Frame = +2
Query: 9 IEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNG----VRCI 62
I + + VTGG G +GS L L+ RG + V A +L D + + +G +R
Sbjct: 2 IRHQVVCVTGGSGCIGSWLVHLLLDRG-YTVHATVQNLNDEAETKHLQSLDGASTRLRLF 178
Query: 63 QGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIK 122
Q D++R + V +A+RG VFHLA+ + + + I GT +VL A G++
Sbjct: 179 QMDLLRHDTVLAAVRGCAGVFHLASPCIVDQVHDPQKELLDPAIKGTMNVLTAAKEAGVR 358
Query: 123 RLVYCSTYNVV 133
R+V S+ + V
Sbjct: 359 RVVLTSSISAV 391
>TC225292 weakly similar to GB|AAM64538.1|21592589|AY086975 cinnamoyl-CoA
reductase-like protein {Arabidopsis thaliana;} , partial
(87%)
Length = 1195
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/137 (25%), Positives = 68/137 (49%), Gaps = 6/137 (4%)
Frame = +3
Query: 3 LSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNGVR 60
L++ K + VTG G +GS + L L++RG + V A D++D + + + G +
Sbjct: 27 LNKQSNPMSKVVCVTGASGAIGSWVALLLLQRG-YTVHATVQDIKDENETKHLEEMEGAK 203
Query: 61 C----IQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDAC 116
+ D++ + + +A++G V HLA + G ++ + I GT +VL A
Sbjct: 204 SRLHFFEMDLLDIDSIAAAIKGCSGVIHLACPNIIGHVEDPEKQILEPAIKGTVNVLKAA 383
Query: 117 LHLGIKRLVYCSTYNVV 133
G++R+V S+ + +
Sbjct: 384 KEAGVERVVATSSISSI 434
>TC210404 similar to UP|Q8H1H5 (Q8H1H5) Cinnamoyl CoA reductase 2 (Fragment)
, partial (55%)
Length = 475
Score = 43.5 bits (101), Expect = 2e-04
Identities = 38/126 (30%), Positives = 61/126 (48%), Gaps = 8/126 (6%)
Frame = +2
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSP----WFAILKDNG----VRCIQ 63
K + VTG GFV S L L+ +G + +RD P + +LK +G + +
Sbjct: 41 KKVCVTGAGGFVASWLVKLLLSKG--YIVHGTVRDPEPATQKYEHLLKLHGASENLTLFK 214
Query: 64 GDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKR 123
D++ E + SA+ G VFHLA S + + + GT +VL+A L ++R
Sbjct: 215 ADLLNYESLRSAISGCTAVFHLACPVPSISVPNPQVEMIEPAVKGTTNVLEASLEAKVQR 394
Query: 124 LVYCST 129
LV+ S+
Sbjct: 395 LVFVSS 412
>TC225293 weakly similar to GB|AAM64538.1|21592589|AY086975 cinnamoyl-CoA
reductase-like protein {Arabidopsis thaliana;} , partial
(97%)
Length = 1421
Score = 43.5 bits (101), Expect = 2e-04
Identities = 34/128 (26%), Positives = 66/128 (51%), Gaps = 6/128 (4%)
Frame = +1
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNGVRC----IQGD 65
K + VTG G +GS + L L++RG + V A D++D + + + G + + D
Sbjct: 136 KVVCVTGASGAIGSWVVLLLLQRG-YTVHATVQDIKDENETKHLEEMEGAKSHLHFFEMD 312
Query: 66 IVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLV 125
++ + + +A++G V HLA + G+ ++ + I GT +VL A G++R+V
Sbjct: 313 LLDIDSIAAAIKGCSGVIHLACPNIIGQVEDPEKQILEPAIKGTVNVLKAAKEAGVERVV 492
Query: 126 YCSTYNVV 133
S+ + +
Sbjct: 493 ATSSISSI 516
>TC217827
Length = 1189
Score = 43.1 bits (100), Expect = 3e-04
Identities = 37/128 (28%), Positives = 65/128 (49%), Gaps = 7/128 (5%)
Frame = +1
Query: 9 IEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNG----VRCI 62
+E VTGG GF+ S L L+ +G H VR + D + + +G ++ +
Sbjct: 31 LEMPEFCVTGGTGFIASYLVKALLEKG-HTVRTTVRNPGDVEKVGFLTELSGAKERLKIL 207
Query: 63 QGDIVRKEDVESALRGADCVFHLAA-FGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGI 121
+ D++ + + A+RG D VFH+A+ + E +Q +D I GT +VL++C+ +
Sbjct: 208 KADLLVEGSFDEAVRGVDGVFHMASPVLIPYDENVQQNLIDPC-IKGTLNVLNSCVKATV 384
Query: 122 KRLVYCST 129
K V S+
Sbjct: 385 KHFVLTSS 408
>TC204029 homologue to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (92%)
Length = 1640
Score = 43.1 bits (100), Expect = 3e-04
Identities = 36/123 (29%), Positives = 58/123 (46%), Gaps = 5/123 (4%)
Frame = +2
Query: 8 GIEGKT--LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCI--- 62
GI G+ +VVTGG GFVGS L +L+ RG D+ +F K+N V
Sbjct: 389 GIGGRRQRIVVTGGAGFVGSHLVDKLIARGD------DVIVIDNFFTGRKENLVHLFGNP 550
Query: 63 QGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIK 122
+ +++R + VE L D ++HLA + + N+ GT ++L +G +
Sbjct: 551 RFELIRHDVVEPILLEVDQIYHLACPASPVHYKYNPVKTIKTNVMGTLNMLGLAKRIGAR 730
Query: 123 RLV 125
L+
Sbjct: 731 FLL 739
>TC218842 similar to UP|Q9SPJ5 (Q9SPJ5) Dihydroflavonol-4-reductase DFR1,
partial (42%)
Length = 496
Score = 42.0 bits (97), Expect = 6e-04
Identities = 34/129 (26%), Positives = 65/129 (50%), Gaps = 7/129 (5%)
Frame = +2
Query: 8 GIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRA-----FDLRDSSPWFAIL-KDNGVRC 61
G + +T+ VTG G++GS L + L+ RG + VRA D+R+ + ++ +
Sbjct: 62 GSKSETVCVTGASGYIGSWLVMRLIERG-YTVRATVLDPADMREVKHLLDLPGAESKLSL 238
Query: 62 IQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-G 120
+ ++ + + A++G VFHLA + K + + I G +++ ACL
Sbjct: 239 WKAELTEEGSFDEAIKGCTGVFHLAT-PVDFKSKDPENEMIKPTIQGVLNIMKACLKAKT 415
Query: 121 IKRLVYCST 129
++RLV+ S+
Sbjct: 416 VRRLVFTSS 442
>TC214784 similar to UP|Q9LZI2 (Q9LZI2) DTDP-glucose 4-6-dehydratase homolog
D18 (At3g62830/F26K9_260), partial (84%)
Length = 1652
Score = 41.6 bits (96), Expect = 8e-04
Identities = 32/119 (26%), Positives = 54/119 (44%), Gaps = 3/119 (2%)
Frame = +3
Query: 10 EGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCI---QGDI 66
+G +VVTGG GFVGS L L+ RG + +F K+N + + ++
Sbjct: 369 KGLRIVVTGGAGFVGSHLVDRLIARGDSVIVV------DNFFTGRKENVMHHFGNPRFEL 530
Query: 67 VRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLV 125
+R + VE L D ++HLA + + N+ GT ++L +G + L+
Sbjct: 531 IRHDVVEPLLLEVDQIYHLACPASPVHYKFNPVKTIKTNVVGTLNMLGLAKRVGARFLL 707
>TC217923 UP|Q9SPJ5 (Q9SPJ5) Dihydroflavonol-4-reductase DFR1, complete
Length = 1254
Score = 40.8 bits (94), Expect = 0.001
Identities = 37/137 (27%), Positives = 65/137 (47%), Gaps = 11/137 (8%)
Frame = +3
Query: 8 GIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILK-------DNGVR 60
G +++ VTG GF+GS L + L++RG + VRA +RD + + +
Sbjct: 54 GSASESVCVTGASGFIGSWLVMRLIKRG-YTVRA-TVRDPVNMKKVKNLVELPGAKSKLS 227
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL- 119
+ D+ + + A++G VFH+A M + V + I G ++ ACL
Sbjct: 228 LWKADLAEEGSFDEAIKGCTGVFHVAT-PMDFESKDPENEVIKPTINGVLDIMKACLKAK 404
Query: 120 GIKRLVYCS---TYNVV 133
++RL++ S T NV+
Sbjct: 405 TVRRLIFTSSAGTLNVI 455
>TC222214 weakly similar to UP|Q89HR7 (Q89HR7) Bll5923 protein , partial (9%)
Length = 411
Score = 40.4 bits (93), Expect = 0.002
Identities = 38/125 (30%), Positives = 61/125 (48%), Gaps = 1/125 (0%)
Frame = +3
Query: 7 EGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDI 66
E E ++VTG GF+G LC LVR+G + VR +R +S A+ + GDI
Sbjct: 6 EAEETMKILVTGASGFLGGKLCDALVRQG-YSVRVL-VRSTSDISAL--SPHIEIFYGDI 173
Query: 67 VRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-GIKRLV 125
+ +A VFHLAA + + ++ VN+ G +VL A +++L+
Sbjct: 174 TDYASLLAACFSCTLVFHLAA--LVEPWLPDPSKFFSVNVGGLKNVLAAAKETRTVEKLL 347
Query: 126 YCSTY 130
Y S++
Sbjct: 348 YTSSF 362
>TC210161 UP|Q9SDZ2 (Q9SDZ2) 2'-hydroxy isoflavone/dihydroflavonol reductase
homolog (Fragment), complete
Length = 1336
Score = 40.4 bits (93), Expect = 0.002
Identities = 61/256 (23%), Positives = 108/256 (41%), Gaps = 16/256 (6%)
Frame = +1
Query: 16 VTGGLGFVGSALCLELVRRGAHEVRAF-----DLRDSSPWFAIL--KDNGVRCIQGDIVR 68
VTGG GF+GS + L+ G + V + R + L ++ + D+
Sbjct: 31 VTGGTGFIGSWIIKRLLEDG-YSVNTTVRPDPEHRKDVSFLTSLPRASQRLQILSADLSN 207
Query: 69 KEDVESALRGADCVFHLAA---FGMSGKEMLQLTRVDQVNITGTCHVLDACLH-LGIKRL 124
E +++ G VFH+A F + E + V + +I G +L ACL+ +KR+
Sbjct: 208 PESFIASIEGCMGVFHVATPVDFELREPEEV----VTKRSIEGALGILKACLNSKTVKRV 375
Query: 125 VYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNC 184
VY S+ + V + + S P+G S S+++ L K + N
Sbjct: 376 VYTSSASAVDNNKEEIMDESSWNDVDYLRSSKPFGWSYSVSKTLTEK---AVLEFGEQNG 546
Query: 185 LYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQ-----TVKSDWVFVENLVLAL 239
L + P ++GP LP + + L F +G++ +++D V V+++ A
Sbjct: 547 LDVVTLIPTLVFGPFICPKLPSSVRNS----LDFILGEKGTFGVVLQTDMVHVDDVARAH 714
Query: 240 ILASMGLLDDSAGKGK 255
I LL+ KG+
Sbjct: 715 IF----LLEHPNPKGR 750
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,912,253
Number of Sequences: 63676
Number of extensions: 343525
Number of successful extensions: 2162
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 2145
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2160
length of query: 479
length of database: 12,639,632
effective HSP length: 101
effective length of query: 378
effective length of database: 6,208,356
effective search space: 2346758568
effective search space used: 2346758568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0335a.3


