

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0334a.8
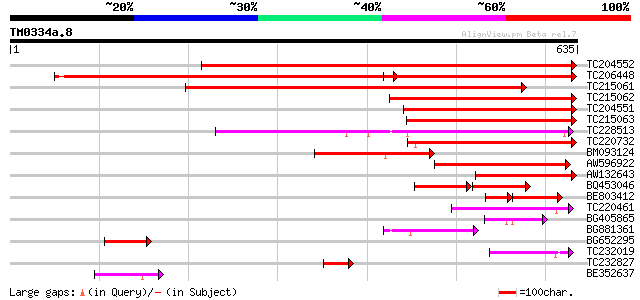
(635 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204552 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic pr... 722 0.0
TC206448 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent... 672 0.0
TC215061 similar to UP|O24550 (O24550) Malate dehydrogenase , p... 650 0.0
TC215062 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic ... 353 1e-97
TC204551 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic pr... 325 4e-89
TC215063 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic ... 317 1e-86
TC228513 similar to UP|MAOM_SOLTU (P37221) NAD-dependent malic e... 308 6e-84
TC220732 similar to UP|Q6PMI1 (Q6PMI1) NADP-dependent malic enzy... 304 9e-83
BM093124 247 1e-65
AW596922 similar to GP|8118507|gb| NADP-dependent malic protein ... 183 2e-46
AW132643 similar to GP|8118507|gb| NADP-dependent malic protein ... 182 5e-46
BQ453046 similar to GP|8894554|emb NAD-dependent malic enzyme (m... 65 4e-23
BE803412 98 1e-20
TC220461 similar to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enzym... 97 3e-20
BG405865 homologue to GP|8118507|gb|A NADP-dependent malic prote... 80 4e-15
BG881361 homologue to GP|8894554|emb NAD-dependent malic enzyme ... 78 1e-14
BG652295 similar to SP|P51615|MAOX_ NADP-dependent malic enzyme ... 70 4e-12
TC232019 homologue to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enz... 69 5e-12
TC232827 similar to UP|Q94G60 (Q94G60) Sucrose synthase , parti... 55 1e-07
BE352637 similar to SP|P37225|MAON NAD-dependent malic enzyme 59... 51 2e-06
>TC204552 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic protein ,
partial (66%)
Length = 1640
Score = 722 bits (1863), Expect = 0.0
Identities = 354/419 (84%), Positives = 389/419 (92%)
Frame = +3
Query: 216 EVLKNWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTID 275
EVL+NWPEK IQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPS+CLP+TID
Sbjct: 3 EVLRNWPEKNIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSACLPITID 182
Query: 276 VGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFN 335
VGTNNE LLNDE Y GL+Q+RATG+EY EL+ EFM AVKQ YGEKVLIQFEDFANHNAF+
Sbjct: 183 VGTNNEKLLNDELYIGLKQRRATGQEYAELMHEFMTAVKQTYGEKVLIQFEDFANHNAFD 362
Query: 336 LLDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIA 395
LL++Y S+HLVFNDDIQGTASVVLAGL+ASLKLVGG LADH FLFLGAGEAGTGIAELIA
Sbjct: 363 LLEKYRSTHLVFNDDIQGTASVVLAGLVASLKLVGGNLADHRFLFLGAGEAGTGIAELIA 542
Query: 396 LEISKQTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIK 455
LE SKQT AP+EE RK +WLVDSKGLIV SR +SLQHFKKPWAHEHEPVK LLDAV IK
Sbjct: 543 LETSKQTNAPLEEVRKNIWLVDSKGLIVSSRKDSLQHFKKPWAHEHEPVKNLLDAVNKIK 722
Query: 456 PTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFAS 515
PTVLIG+SG G+TFTKEV+EAMAS+NE+P+IL+LSNPTSQSECTAEEAY WS+G+AIFAS
Sbjct: 723 PTVLIGTSGQGRTFTKEVIEAMASINERPIILSLSNPTSQSECTAEEAYKWSQGRAIFAS 902
Query: 516 GSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQEN 575
GSPF PV+Y GKV+ PGQANNAYIFPGFGLGLI+SG IRV D++LLAASEALAAQVSQEN
Sbjct: 903 GSPFPPVEYEGKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLLAASEALAAQVSQEN 1082
Query: 576 YDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
+DKGLIYPPFTNIRKISAHIAA+VAAK YELGLA+ LP+PKDLVK+AESCMY+P YRSY
Sbjct: 1083FDKGLIYPPFTNIRKISAHIAANVAAKAYELGLATRLPQPKDLVKFAESCMYTPSYRSY 1259
>TC206448 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent malic
enzyme {Phaseolus vulgaris;} , partial (97%)
Length = 2208
Score = 672 bits (1734), Expect = 0.0
Identities = 334/385 (86%), Positives = 355/385 (91%)
Frame = +3
Query: 51 RNGGEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLA 110
RNGGE D+ N GGVRDLYGED ATEDQLITPWTFSVASG +LLRDPRYNKGLA
Sbjct: 123 RNGGE-----DYSNGHGGGGVRDLYGEDCATEDQLITPWTFSVASGCSLLRDPRYNKGLA 287
Query: 111 FTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKI 170
FTE ERDAHYLRGLLPPA+F QELQEKR MHN+RQYEVPLH+YMAMMDLQERNERLFYK+
Sbjct: 288 FTEGERDAHYLRGLLPPAIFNQELQEKRLMHNLRQYEVPLHRYMAMMDLQERNERLFYKL 467
Query: 171 LIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIV 230
LI+NVEELLP+VYTP VGEACQKYGSI++RPQGLYISLKEKGKILEVLKNWPEK+IQVIV
Sbjct: 468 LINNVEELLPVVYTPTVGEACQKYGSIFRRPQGLYISLKEKGKILEVLKNWPEKSIQVIV 647
Query: 231 VTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYT 290
VTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLP+TIDVGTNNE LLNDEFY
Sbjct: 648 VTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPITIDVGTNNEKLLNDEFYI 827
Query: 291 GLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDD 350
GL+QKRATG+EY ELL+EFM AVKQNYGEKVLIQFEDFANHNAF+LL++YSSSHLVFNDD
Sbjct: 828 GLKQKRATGQEYAELLEEFMHAVKQNYGEKVLIQFEDFANHNAFDLLEKYSSSHLVFNDD 1007
Query: 351 IQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETR 410
IQGTASVVLAGLLASLKLVGGTL DHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETR
Sbjct: 1008IQGTASVVLAGLLASLKLVGGTLVDHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETR 1187
Query: 411 KKVWLVDSKGLIVRSRLESLQHFKK 435
KK+WLVDSKG I SR + +K
Sbjct: 1188KKIWLVDSKGKIASSRSRTTSPVQK 1262
Score = 405 bits (1042), Expect = e-113
Identities = 202/216 (93%), Positives = 208/216 (95%)
Frame = +2
Query: 419 KGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMA 478
KGLIV SRLESLQ FKKPWAHEHEPVK+LLDAVKAIKPTVLIGSSG GKTFTKEVVE MA
Sbjct: 1313 KGLIVSSRLESLQQFKKPWAHEHEPVKSLLDAVKAIKPTVLIGSSGAGKTFTKEVVETMA 1492
Query: 479 SLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAY 538
SLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPV+Y GK++ PGQANNAY
Sbjct: 1493 SLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVEYEGKLFVPGQANNAY 1672
Query: 539 IFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAAS 598
IFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPF+NIRKISAHIAA
Sbjct: 1673 IFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFSNIRKISAHIAAK 1852
Query: 599 VAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
VAAK Y+LGLASHLPRPKDLVKYAESCMYSPGYRSY
Sbjct: 1853 VAAKAYDLGLASHLPRPKDLVKYAESCMYSPGYRSY 1960
>TC215061 similar to UP|O24550 (O24550) Malate dehydrogenase , partial (58%)
Length = 1146
Score = 650 bits (1677), Expect = 0.0
Identities = 320/381 (83%), Positives = 352/381 (91%)
Frame = +1
Query: 198 YKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSL 257
+KRPQGL+ISLKEKGK+LEVLKNWPEK+IQVIVVTDGERILGLGDLGCQGMGIPVGKL+L
Sbjct: 1 FKRPQGLFISLKEKGKVLEVLKNWPEKSIQVIVVTDGERILGLGDLGCQGMGIPVGKLAL 180
Query: 258 YTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNY 317
YTALGGVRPS+CLP+TIDVGTNNE LLNDEFY GLRQKRATG+EY ELL EFM AVKQNY
Sbjct: 181 YTALGGVRPSACLPITIDVGTNNEKLLNDEFYIGLRQKRATGQEYSELLQEFMTAVKQNY 360
Query: 318 GEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHT 377
GEKVLIQFEDFANHNAF LL +Y ++HLVFNDDIQGTASVVLAG++A+LKL+GG LADHT
Sbjct: 361 GEKVLIQFEDFANHNAFELLAKYGTTHLVFNDDIQGTASVVLAGVVAALKLIGGNLADHT 540
Query: 378 FLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPW 437
FLFLGAGEAGTGIAELIALE+SKQTK P+EETRKK+WLVDSKGLIV SR SLQHFK+PW
Sbjct: 541 FLFLGAGEAGTGIAELIALEMSKQTKTPIEETRKKIWLVDSKGLIVGSRKASLQHFKQPW 720
Query: 438 AHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSE 497
AHEHEPV +LL+AVK IKPTVLIGSSGVGKTFTKEV+EA+ S+NEKPL+LALSNPTSQSE
Sbjct: 721 AHEHEPVGSLLEAVKVIKPTVLIGSSGVGKTFTKEVIEAVTSINEKPLVLALSNPTSQSE 900
Query: 498 CTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRD 557
CTAEEAY WS+G+AIFASGSPFDPV+Y GKVY GQANNAYIFPGFGLGL+ISGAIRV D
Sbjct: 901 CTAEEAYQWSEGRAIFASGSPFDPVEYKGKVYYSGQANNAYIFPGFGLGLVISGAIRVHD 1080
Query: 558 EMLLAASEALAAQVSQENYDK 578
+ML AS ALA V E Y++
Sbjct: 1081DMLSTASVALAKLVYNEYYEQ 1143
>TC215062 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic enzyme
(NADP-ME) , partial (36%)
Length = 971
Score = 353 bits (907), Expect = 1e-97
Identities = 172/209 (82%), Positives = 194/209 (92%)
Frame = +2
Query: 426 RLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPL 485
R SLQHFK+PWAHEHEPV +LL+AVK IKPTVLIGSSGVGKTFTKEV+EA+ S+NEKPL
Sbjct: 2 RKASLQHFKQPWAHEHEPVGSLLEAVKVIKPTVLIGSSGVGKTFTKEVIEAVTSINEKPL 181
Query: 486 ILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGL 545
+LALSNPTSQSECTAEEAY WS+G+AIFASGSPFDPV+Y GKVY GQANNAYIFPGFGL
Sbjct: 182 VLALSNPTSQSECTAEEAYEWSEGRAIFASGSPFDPVEYKGKVYYSGQANNAYIFPGFGL 361
Query: 546 GLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYE 605
GL+ISGAIRV D+MLLAASEALA V++ENY+KGLIYPPF+NIRKISA+IAASVAAK YE
Sbjct: 362 GLVISGAIRVHDDMLLAASEALAKLVTEENYEKGLIYPPFSNIRKISANIAASVAAKAYE 541
Query: 606 LGLASHLPRPKDLVKYAESCMYSPGYRSY 634
LGLA+ LPRP++LVKYAESCMY+P YR+Y
Sbjct: 542 LGLATRLPRPQNLVKYAESCMYTPVYRNY 628
>TC204551 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic protein ,
partial (30%)
Length = 856
Score = 325 bits (833), Expect = 4e-89
Identities = 158/193 (81%), Positives = 179/193 (91%)
Frame = +2
Query: 442 EPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAE 501
EPVK LLDAV IKPT LIG+SG G+TFTKEV+EAMAS+N++P+IL+LSNPTSQSECTAE
Sbjct: 2 EPVKNLLDAVNKIKPTELIGTSGQGRTFTKEVIEAMASINKRPIILSLSNPTSQSECTAE 181
Query: 502 EAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLL 561
EAY WS+G+AIFASGSPF PV+Y GKV+ PGQANNAYIFPGFGLGLI+SG IRV D++LL
Sbjct: 182 EAYKWSQGRAIFASGSPFPPVEYEGKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLL 361
Query: 562 AASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKY 621
AASEALAAQVSQEN+DKGLIYPPFTNIRKISAHIAA+VAAK YELGLA+ LP+PKDLVK+
Sbjct: 362 AASEALAAQVSQENFDKGLIYPPFTNIRKISAHIAANVAAKAYELGLATRLPQPKDLVKF 541
Query: 622 AESCMYSPGYRSY 634
AESCMY+P YRSY
Sbjct: 542 AESCMYTPAYRSY 580
>TC215063 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic enzyme
(NADP-ME) , partial (32%)
Length = 918
Score = 317 bits (812), Expect = 1e-86
Identities = 156/190 (82%), Positives = 176/190 (92%)
Frame = +2
Query: 445 KALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAY 504
++LL+AVK IKPTVLIGSSGVGKTFTKEV+EA+ S+NEKPL+LALSNPTSQSECTAEEAY
Sbjct: 5 ESLLEAVKVIKPTVLIGSSGVGKTFTKEVIEAVTSINEKPLVLALSNPTSQSECTAEEAY 184
Query: 505 TWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAAS 564
WS+G+AIFASGSPFDPV+Y GKVY GQANNAYIFPGFGLGL+ISGAIRV D+MLLAAS
Sbjct: 185 QWSEGRAIFASGSPFDPVEYKGKVYYSGQANNAYIFPGFGLGLVISGAIRVHDDMLLAAS 364
Query: 565 EALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAES 624
EALA VS ENY+KGLIYPPF+NIR+ISA+IAASVA K YELGLA+ LPRP++LVKYAES
Sbjct: 365 EALAKLVSNENYEKGLIYPPFSNIREISANIAASVAGKAYELGLATRLPRPQNLVKYAES 544
Query: 625 CMYSPGYRSY 634
CMYSP YR+Y
Sbjct: 545 CMYSPVYRNY 574
>TC228513 similar to UP|MAOM_SOLTU (P37221) NAD-dependent malic enzyme 62 kDa
isoform, mitochondrial precursor (NAD-ME) , partial
(69%)
Length = 1451
Score = 308 bits (788), Expect = 6e-84
Identities = 183/427 (42%), Positives = 253/427 (58%), Gaps = 26/427 (6%)
Frame = +2
Query: 231 VTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYT 290
VTDG RILGLGDLG QG+GI +GKL LY A G+ P LPV IDVGTNNE LL D Y
Sbjct: 2 VTDGSRILGLGDLGVQGIGIAIGKLDLYVAAAGINPQRVLPVMIDVGTNNEKLLEDPLYL 181
Query: 291 GLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDD 350
GL+Q R G +Y ++DEFM AV + V++QFEDF + AF LL RY +++ +FNDD
Sbjct: 182 GLQQHRLDGDDYLAVVDEFMEAVFTRW-PNVIVQFEDFQSKWAFKLLQRYRNTYRMFNDD 358
Query: 351 IQGTASVVLAGLLASLKLVGGTLAD---HTFLFLGAGEAGTGIAELIALEISK---QTKA 404
+QGTA V +AGLL +++ G + D + GAG AG G+ +++ +
Sbjct: 359 VQGTAGVAIAGLLGAVRAQGRPMIDFPKQKIVVAGAGSAGIGVLNAARKTMARMLGNNEV 538
Query: 405 PVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPV--------KALLDAVKAIKP 456
E + + W+VD++GLI R E++ P+A + + +L++ VK +KP
Sbjct: 539 AFESAKSQFWVVDAQGLITEGR-ENIDPDALPFARNLKEMDRQGLREGASLVEVVKQVKP 715
Query: 457 TVLIGSSGVGKTFTKEVVEAM-ASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFAS 515
VL+G S VG F+KEV+EA+ S + +P I A+SNPT +ECTAEEA++ IFAS
Sbjct: 716 DVLLGLSAVGGLFSKEVLEALKGSTSTRPAIFAMSNPTKNAECTAEEAFSILGDNIIFAS 895
Query: 516 GSPFDPVKYG-GKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQE 574
GSPF V G G + Q NN Y+FPG GLG ++SGA V D ML AA+E LA +S+E
Sbjct: 896 GSPFSNVDLGNGHIGHCNQGNNMYLFPGIGLGTLLSGARIVSDGMLQAAAERLATYMSEE 1075
Query: 575 NYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLAS--HLPRPKDLVK--------YAES 624
KG+I+P + IR I+ +A +V + E LA H ++L K Y ++
Sbjct: 1076EVLKGIIFPSTSRIRDITKQVATAVIKEAVEEDLAEGYHGMDARELQKLSEDEIAEYVQN 1255
Query: 625 CMYSPGY 631
M+SP Y
Sbjct: 1256NMWSPEY 1276
>TC220732 similar to UP|Q6PMI1 (Q6PMI1) NADP-dependent malic enzyme 3 ,
partial (32%)
Length = 662
Score = 304 bits (778), Expect = 9e-83
Identities = 154/193 (79%), Positives = 173/193 (88%), Gaps = 4/193 (2%)
Frame = +1
Query: 446 ALLDAVKA----IKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAE 501
ALL + A IKPTVLIGSSGVG+TFTKEVVEAM S N+KPLILALSNPTSQSECTAE
Sbjct: 4 ALLSLITAHFQVIKPTVLIGSSGVGRTFTKEVVEAMTSNNDKPLILALSNPTSQSECTAE 183
Query: 502 EAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLL 561
EAY WS+G+AIFASGSPFDPV+Y GKVYA GQANNAYIFPGFGLGL+ISGAIRV D+MLL
Sbjct: 184 EAYQWSEGRAIFASGSPFDPVEYKGKVYASGQANNAYIFPGFGLGLVISGAIRVHDDMLL 363
Query: 562 AASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKY 621
AASE+LA QVS+ENY GLIYPPF+NIR+ISA+IAA+VAAK YELGLA+ LPRP++LVK
Sbjct: 364 AASESLAKQVSEENYKNGLIYPPFSNIRRISANIAANVAAKAYELGLATRLPRPQNLVKC 543
Query: 622 AESCMYSPGYRSY 634
AESCMY+P YR+Y
Sbjct: 544 AESCMYTPVYRNY 582
>BM093124
Length = 420
Score = 247 bits (631), Expect = 1e-65
Identities = 129/139 (92%), Positives = 131/139 (93%), Gaps = 5/139 (3%)
Frame = +2
Query: 342 SSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQ 401
SSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQ
Sbjct: 2 SSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQ 181
Query: 402 TKAPVEETRKKVWLVDSK-----GLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKP 456
TKAPVE+TRKK+WLVDSK GLIV SRLESLQHFKKPWAHEHEPVK LLDAVKAIKP
Sbjct: 182 TKAPVEKTRKKIWLVDSKVKMPQGLIVSSRLESLQHFKKPWAHEHEPVKTLLDAVKAIKP 361
Query: 457 TVLIGSSGVGKTFTKEVVE 475
TVLIGSSG GKTFTKEVVE
Sbjct: 362 TVLIGSSGAGKTFTKEVVE 418
>AW596922 similar to GP|8118507|gb| NADP-dependent malic protein {Ricinus
communis}, partial (16%)
Length = 460
Score = 183 bits (464), Expect = 2e-46
Identities = 94/153 (61%), Positives = 116/153 (75%)
Frame = +2
Query: 476 AMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQAN 535
A AS+NE+P+IL+LSNPT QSECTAEEA WS G+AIFASGSPF PV+Y GKV+ PGQAN
Sbjct: 2 ARASINERPIILSLSNPT*QSECTAEEANEWSHGRAIFASGSPFPPVEYEGKVFVPGQAN 181
Query: 536 NAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHI 595
NAYIFPGFGLGLI+SG I V+++ + AA EA AAQ+SQE D+ LI K AH+
Sbjct: 182 NAYIFPGFGLGLIMSGTILVQEDQVEAACEA*AAQLSQETMDQELICRTSKQDEKDVAHV 361
Query: 596 AASVAAKVYELGLASHLPRPKDLVKYAESCMYS 628
+A+ A+ Y+LGL + LP+ KD K AESC Y+
Sbjct: 362 SANGTAEAYDLGLTTELPQAKDR*KVAESCEYA 460
>AW132643 similar to GP|8118507|gb| NADP-dependent malic protein {Ricinus
communis}, partial (17%)
Length = 381
Score = 182 bits (461), Expect = 5e-46
Identities = 88/113 (77%), Positives = 103/113 (90%)
Frame = +3
Query: 522 VKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLI 581
V+Y GKV+ PGQANNAYIFPGFGLGLI+SG IRV D++LLAASEALA+QV+Q++YDKGLI
Sbjct: 6 VEYDGKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLLAASEALASQVTQKDYDKGLI 185
Query: 582 YPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
YPPF+NIRKISA IAA+VAAK YELGLA+ LP+PKDLVK+AES MY+P YR Y
Sbjct: 186 YPPFSNIRKISARIAANVAAKAYELGLATSLPQPKDLVKFAESSMYTPLYRGY 344
>BQ453046 similar to GP|8894554|emb NAD-dependent malic enzyme (malate
oxidoreductase) {Cicer arietinum}, partial (44%)
Length = 407
Score = 64.7 bits (156), Expect(2) = 4e-23
Identities = 31/66 (46%), Positives = 45/66 (67%), Gaps = 1/66 (1%)
Frame = +1
Query: 519 FDPVKYG-GKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYD 577
F+ V G G+ + QANN Y+FPG GLG ++SGA + D ML AA+E LA+ ++ E+
Sbjct: 199 FENVDLGNGEFFHVNQANNMYLFPGIGLGTLLSGARHITDGMLRAAAECLASYMTDEDVQ 378
Query: 578 KGLIYP 583
KG++YP
Sbjct: 379 KGILYP 396
Score = 62.0 bits (149), Expect(2) = 4e-23
Identities = 33/65 (50%), Positives = 44/65 (66%), Gaps = 1/65 (1%)
Frame = +2
Query: 454 IKPTVLIGSSGVGKTFTKEVVEAMA-SLNEKPLILALSNPTSQSECTAEEAYTWSKGKAI 512
+KP VL+G SGVG F EV++AM S++ KP I A+SNPT +ECTA EA++ +
Sbjct: 2 VKPHVLLGLSGVGGVFNAEVLKAMRESVSTKPAIFAMSNPTMNAECTAIEAFSHAGENIA 181
Query: 513 FASGS 517
FA GS
Sbjct: 182 FAIGS 196
>BE803412
Length = 411
Score = 97.8 bits (242), Expect = 1e-20
Identities = 48/56 (85%), Positives = 53/56 (93%)
Frame = +2
Query: 564 SEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLV 619
+EALAAQVSQEN+DKGLIYPPFTNIRKISAHIAA+VAAK YELGLA+ L +PKDLV
Sbjct: 242 AEALAAQVSQENFDKGLIYPPFTNIRKISAHIAANVAAKAYELGLATRLTQPKDLV 409
Score = 53.1 bits (126), Expect = 4e-07
Identities = 24/31 (77%), Positives = 28/31 (89%)
Frame = +1
Query: 533 QANNAYIFPGFGLGLIISGAIRVRDEMLLAA 563
QANNAYIFPGFGLGLI+ G IRV D++L+AA
Sbjct: 4 QANNAYIFPGFGLGLIMYGTIRVHDDLLVAA 96
>TC220461 similar to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enzyme (Malate
oxidoreductase) (Fragment) , partial (59%)
Length = 1101
Score = 96.7 bits (239), Expect = 3e-20
Identities = 58/147 (39%), Positives = 85/147 (57%), Gaps = 11/147 (7%)
Frame = +1
Query: 496 SECTAEEAYTWSKGKAIFASGSPFDPVKYG-GKVYAPGQANNAYIFPGFGLGLIISGAIR 554
+ECTA EA++ + +FASGSPF+ V G G+V QANN Y+FPG GLG ++SGA
Sbjct: 352 AECTAIEAFSHAGENIVFASGSPFENVDLGNGEVGHVNQANNMYLFPGIGLGTLLSGARH 531
Query: 555 VRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHI-AASVAAKVYELGLASH-- 611
+ D ML AA+E LA+ +++++ KG++YP IR ++A + AA V A V E H
Sbjct: 532 ITDGMLRAAAECLASYMTEDDVRKGILYPSIDCIRDVTAEVGAAVVCAAVAEKQAEGHGD 711
Query: 612 -------LPRPKDLVKYAESCMYSPGY 631
++ V+Y M+ P Y
Sbjct: 712 VGFKELENMSKEETVEYVRGNMWYPEY 792
>BG405865 homologue to GP|8118507|gb|A NADP-dependent malic protein {Ricinus
communis}, partial (11%)
Length = 352
Score = 79.7 bits (195), Expect = 4e-15
Identities = 53/115 (46%), Positives = 59/115 (51%), Gaps = 44/115 (38%)
Frame = +1
Query: 532 GQANNAYIFPGFGLGLIISGAIRV--------------------------------RDEM 559
GQANNAYIFPGFGL G R+E+
Sbjct: 4 GQANNAYIFPGFGLV**CLGPFECMMTCFWLPVSAQDTCEH*FQTYVWDR*K*DHGRNEI 183
Query: 560 LL------------AASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAK 602
+ +EALAAQVSQEN+DKGLIYPPFTNIRKISAHIAA+VAAK
Sbjct: 184 YIWN*YNVILFDXDGTAEALAAQVSQENFDKGLIYPPFTNIRKISAHIAANVAAK 348
>BG881361 homologue to GP|8894554|emb NAD-dependent malic enzyme (malate
oxidoreductase) {Cicer arietinum}, partial (44%)
Length = 412
Score = 77.8 bits (190), Expect = 1e-14
Identities = 46/114 (40%), Positives = 68/114 (59%), Gaps = 7/114 (6%)
Frame = +2
Query: 419 KGLIVRSRLESLQHFKKPWAHEHEPVKAL------LDAVKAIKPTVLIGSSGVGKTFTKE 472
+GL+ R SL P+A ++ L ++ VK I+P VL+G SGVG F +E
Sbjct: 65 EGLVTTER-NSLDPAAAPFAKNPRDIEGLTEGASIIEVVKKIRPHVLLGLSGVGGIFNEE 241
Query: 473 VVEAMA-SLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYG 525
V++AM S++ KP I A+SNPT +ECT+ +A+ + +FASGSPF+ V G
Sbjct: 242 VLKAMRESVSTKPAIFAMSNPTMNAECTSIDAFKHAGENIVFASGSPFENVDLG 403
>BG652295 similar to SP|P51615|MAOX_ NADP-dependent malic enzyme (EC
1.1.1.40) (NADP-ME). [Grape] {Vitis vinifera}, partial
(8%)
Length = 410
Score = 69.7 bits (169), Expect = 4e-12
Identities = 34/53 (64%), Positives = 40/53 (75%)
Frame = +2
Query: 107 KGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDL 159
K LAF KERDAH L GLLPP +Q+LQE + M+NI Y+VPL KY+AMMDL
Sbjct: 164 KELAFLTKERDAHSLCGLLPPTFSSQQLQENKLMNNISLYQVPLQKYVAMMDL 322
>TC232019 homologue to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enzyme (Malate
oxidoreductase) (Fragment) , partial (37%)
Length = 616
Score = 69.3 bits (168), Expect = 5e-12
Identities = 40/105 (38%), Positives = 61/105 (58%), Gaps = 11/105 (10%)
Frame = +2
Query: 538 YIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAA 597
Y+FPG GLG ++SGA + D ML AASE LA+ +++E+ KG++YP +IR ++A + A
Sbjct: 5 YLFPGIGLGTLLSGAHLITDGMLQAASECLASYMAEEDILKGILYPSVDSIRDVTAEVGA 184
Query: 598 SVAAKVYELGLA-----------SHLPRPKDLVKYAESCMYSPGY 631
+V E LA SH+ + + V+Y S M+ P Y
Sbjct: 185 AVLRAAVEEELAEGHGDVGPKELSHMSK-DEAVEYVRSNMWYPVY 316
>TC232827 similar to UP|Q94G60 (Q94G60) Sucrose synthase , partial (8%)
Length = 899
Score = 54.7 bits (130), Expect = 1e-07
Identities = 27/34 (79%), Positives = 29/34 (84%)
Frame = +2
Query: 352 QGTASVVLAGLLASLKLVGGTLADHTFLFLGAGE 385
QGT SVVLAGL+A+LKLVGG L DH FLFLGA E
Sbjct: 503 QGTTSVVLAGLVAALKLVGGDLTDHMFLFLGARE 604
Score = 30.8 bits (68), Expect = 1.9
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = +2
Query: 385 EAGTGIAELIALEISKQTK 403
+AGTGIAELIALE SK+ +
Sbjct: 812 QAGTGIAELIALETSKRVR 868
>BE352637 similar to SP|P37225|MAON NAD-dependent malic enzyme 59 kDa isoform
mitochondrial precursor (EC 1.1.1.39) (NAD-ME).
[Potato], partial (18%)
Length = 557
Score = 50.8 bits (120), Expect = 2e-06
Identities = 33/87 (37%), Positives = 42/87 (47%), Gaps = 10/87 (11%)
Frame = +2
Query: 96 GNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYE-------- 147
G +L DP +NK F ERD LRGLLPP V + E Q R M++ R E
Sbjct: 149 GADILHDPWFNKDTGFPLTERDRLGLRGLLPPXVISFEQQYDRFMNSFRSLENNTQGLPD 328
Query: 148 --VPLHKYMAMMDLQERNERLFYKILI 172
V + D RNE L+Y++LI
Sbjct: 329 KVVSWQNGGS*XDCMTRNETLYYRVLI 409
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,372,042
Number of Sequences: 63676
Number of extensions: 329908
Number of successful extensions: 1502
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 1477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1492
length of query: 635
length of database: 12,639,632
effective HSP length: 103
effective length of query: 532
effective length of database: 6,081,004
effective search space: 3235094128
effective search space used: 3235094128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0334a.8


