

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
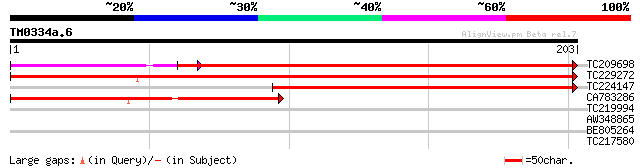
Query= TM0334a.6
(203 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209698 weakly similar to UP|Q9LS52 (Q9LS52) Gb|AAD39678.1, par... 254 2e-68
TC229272 weakly similar to UP|Q9XI47 (Q9XI47) F9L1.16, partial (... 176 5e-45
TC224147 similar to UP|Q9LS52 (Q9LS52) Gb|AAD39678.1, partial (37%) 145 1e-35
CA783286 124 3e-29
TC219994 28 3.7
AW348865 similar to SP|P49730|RIR2 Ribonucleoside-diphosphate re... 27 4.8
BE805264 27 8.3
TC217580 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (74%) 27 8.3
>TC209698 weakly similar to UP|Q9LS52 (Q9LS52) Gb|AAD39678.1, partial (41%)
Length = 1050
Score = 254 bits (649), Expect = 2e-68
Identities = 115/143 (80%), Positives = 132/143 (91%)
Frame = +1
Query: 61 KCKEGETIRDPSQLEFEARSTKDGAWYDVEAFLAHRFVGTGEAEVRVRFVGFGASEDEWV 120
K GET++DPS+LEFEARS+KDGAWYDVEAFLAHRF+ TGEAEV+VRFVGFGA EDEW+
Sbjct: 250 KMDRGETMQDPSELEFEARSSKDGAWYDVEAFLAHRFLNTGEAEVQVRFVGFGAEEDEWI 429
Query: 121 NIKDSVRERSVPFESTDCSYLNVGDPVLCFQERRDQAIYYDARILEIQRRMHDIRGCRCL 180
NIK SVR+RS+P EST+CS L +GDPVLCFQERRDQAIYYDA I+EIQ+RMHDIRGCRCL
Sbjct: 430 NIKTSVRQRSIPLESTECSNLKIGDPVLCFQERRDQAIYYDAHIVEIQKRMHDIRGCRCL 609
Query: 181 ILVRYDHDNTEEKVRLRRLCRRP 203
+L+ YDHDN+EE+VRLRRLCRRP
Sbjct: 610 LLIHYDHDNSEERVRLRRLCRRP 678
Score = 60.8 bits (146), Expect = 4e-10
Identities = 33/69 (47%), Positives = 41/69 (58%)
Frame = +2
Query: 1 MDKLSGESQGRSFDREFYQKLTASFNRSSGRAGKPTVKWTEARIQDLPEVPENNLESSQG 60
M+KL E G S REFYQKL SFN SSGRAGKP +KWTE ++ + NL+
Sbjct: 140 MEKLLREPTGGSLGREFYQKLARSFNYSSGRAGKPIIKWTEEKLCKTHQ--SWNLKQDHQ 313
Query: 61 KCKEGETIR 69
K + G +R
Sbjct: 314 KMEHGMMLR 340
>TC229272 weakly similar to UP|Q9XI47 (Q9XI47) F9L1.16, partial (53%)
Length = 1079
Score = 176 bits (447), Expect = 5e-45
Identities = 88/219 (40%), Positives = 132/219 (60%), Gaps = 16/219 (7%)
Frame = +3
Query: 1 MDKLSGESQGRSFDREFYQKLTASFNRSSGRAGKPTVKWTEARI---------------- 44
++++ + G+ F+R+ Q++ F+ SS AGK ++ W + ++
Sbjct: 156 LERIYKDVVGKVFNRKLCQEIAKRFSSSSNGAGKNSLSWQQVQLWFRNSQRMLLGEDISS 335
Query: 45 QDLPEVPENNLESSQGKCKEGETIRDPSQLEFEARSTKDGAWYDVEAFLAHRFVGTGEAE 104
DL ++ + +S +G+ D + FEARSTKD AW+DV FL +R + TGE E
Sbjct: 336 SDLLKISADLADSPLLGNGKGKQATDLDDMGFEARSTKDNAWHDVSMFLNYRVLSTGELE 515
Query: 105 VRVRFVGFGASEDEWVNIKDSVRERSVPFESTDCSYLNVGDPVLCFQERRDQAIYYDARI 164
VRVR+ GFG +DEW+N+K VRERS+P E ++C + GD VLCF E+ D A+Y DARI
Sbjct: 516 VRVRYAGFGKEQDEWMNVKLGVRERSIPLEPSECHKVKDGDLVLCFLEKEDYALYCDARI 695
Query: 165 LEIQRRMHDIRGCRCLILVRYDHDNTEEKVRLRRLCRRP 203
++IQR++HD C C +V++ HDNTEE V R+C RP
Sbjct: 696 VKIQRKIHDPTDCTCTFIVQFVHDNTEEGVSFSRICCRP 812
>TC224147 similar to UP|Q9LS52 (Q9LS52) Gb|AAD39678.1, partial (37%)
Length = 816
Score = 145 bits (366), Expect = 1e-35
Identities = 63/109 (57%), Positives = 83/109 (75%)
Frame = +3
Query: 95 HRFVGTGEAEVRVRFVGFGASEDEWVNIKDSVRERSVPFESTDCSYLNVGDPVLCFQERR 154
HR++ T + EV RF GFG EDEW+NI+ VR RS+P ES++C + GD +LCFQE +
Sbjct: 3 HRYLETSDPEVLXRFAGFGPEEDEWINIRKHVRPRSLPCESSECVVVIPGDLILCFQEGK 182
Query: 155 DQAIYYDARILEIQRRMHDIRGCRCLILVRYDHDNTEEKVRLRRLCRRP 203
+QA+Y+DA +L+ QRR HD+RGCRC LVRYDHD +EE V LR++CRRP
Sbjct: 183 EQALYFDAHVLDAQRRRHDVRGCRCRFLVRYDHDQSEEIVPLRKICRRP 329
>CA783286
Length = 460
Score = 124 bits (311), Expect = 3e-29
Identities = 65/104 (62%), Positives = 75/104 (71%), Gaps = 6/104 (5%)
Frame = +2
Query: 1 MDKLSGESQGRSFDREFYQKLTASFNRSSGRAGKPTVKWTE------ARIQDLPEVPENN 54
M+KL E G S +EFYQKL SFN SSGRAGKP +KWTE R+QD VP +
Sbjct: 152 MEKLLREPTGGSLGKEFYQKLARSFNYSSGRAGKPIIKWTEIESWFQTRLQDSSLVPSSE 331
Query: 55 LESSQGKCKEGETIRDPSQLEFEARSTKDGAWYDVEAFLAHRFV 98
L KCKEGET++ PS+LEFEARS+KDGAWYDVEAFLAHRF+
Sbjct: 332 LMVP--KCKEGETMQHPSELEFEARSSKDGAWYDVEAFLAHRFL 457
>TC219994
Length = 741
Score = 27.7 bits (60), Expect = 3.7
Identities = 14/50 (28%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Frame = +3
Query: 40 TEARIQDLPEVPENNLESSQ------GKCKEGETIRDPSQLEFEARSTKD 83
++A + +P N + S+ G C + +T+RD L E TKD
Sbjct: 300 SDAMWSQMSHIPSNTVSESRSGSTMLGNCFQHQTMRDAELLNLETARTKD 449
>AW348865 similar to SP|P49730|RIR2 Ribonucleoside-diphosphate reductase
small chain (EC 1.17.4.1) (Ribonucleotide reductase),
partial (60%)
Length = 718
Score = 27.3 bits (59), Expect = 4.8
Identities = 21/79 (26%), Positives = 30/79 (37%)
Frame = +3
Query: 41 EARIQDLPEVPENNLESSQGKCKEGETIRDPSQLEFEARSTKDGAWYDVEAFLAHRFVGT 100
E R+Q+ EV L S EGE P+ LE + R+ G Y ++A H +
Sbjct: 459 EKRVQETREVAVKPLISRDEFVGEGEPRH*PTLLEPKYRAKASGEEYSLDASECHXXLRX 638
Query: 101 GEAEVRVRFVGFGASEDEW 119
+ G D W
Sbjct: 639 XFXXIDPAXXPLGFFGDAW 695
>BE805264
Length = 445
Score = 26.6 bits (57), Expect = 8.3
Identities = 14/43 (32%), Positives = 23/43 (52%), Gaps = 1/43 (2%)
Frame = +2
Query: 35 PTVKWTEAR-IQDLPEVPENNLESSQGKCKEGETIRDPSQLEF 76
P V +E+ +Q + +PE+ LESS G + + DP+ F
Sbjct: 95 PPVNISESEELQPVSIIPESPLESSSGAASAIDAVEDPAFTNF 223
>TC217580 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (74%)
Length = 2139
Score = 26.6 bits (57), Expect = 8.3
Identities = 18/53 (33%), Positives = 25/53 (46%)
Frame = +1
Query: 122 IKDSVRERSVPFESTDCSYLNVGDPVLCFQERRDQAIYYDARILEIQRRMHDI 174
IKD+ + +S S LN+ D L R DQA+ AR L RR ++
Sbjct: 268 IKDARSLIATQDQSEIASALNLVDAALAISPRFDQALELRARALLYLRRFKEV 426
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,295,802
Number of Sequences: 63676
Number of extensions: 101551
Number of successful extensions: 382
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 381
length of query: 203
length of database: 12,639,632
effective HSP length: 93
effective length of query: 110
effective length of database: 6,717,764
effective search space: 738954040
effective search space used: 738954040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0334a.6


