

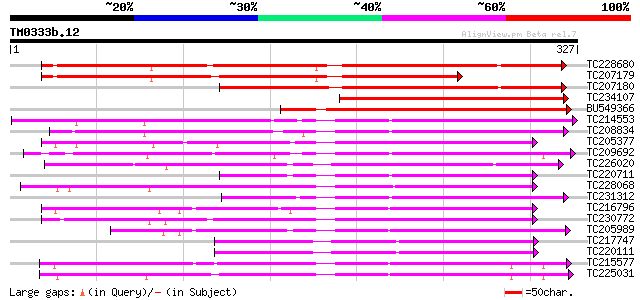
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.12
(327 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228680 similar to UP|G2O1_PEA (Q9SQ80) Gibberellin 2-beta-diox... 288 2e-78
TC207179 similar to UP|G2O1_PEA (Q9SQ80) Gibberellin 2-beta-diox... 244 4e-65
TC207180 similar to GB|AAD45425.1|5579094|AF100955 gibberellin 2... 212 2e-55
TC234107 similar to UP|G2O2_PEA (Q9XHM5) Gibberellin 2-beta-diox... 187 4e-48
BU549366 homologue to GP|4678586|emb GA 2-oxidase {Phaseolus coc... 166 2e-41
TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 152 3e-37
TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 149 2e-36
TC205377 135 3e-32
TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1... 131 4e-31
TC226020 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, ... 128 4e-30
TC220711 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 127 9e-30
TC228068 anthocyanidin synthase [Glycine max] 126 2e-29
TC231312 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1... 126 2e-29
TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {A... 124 4e-29
TC230772 similar to UP|O24648 (O24648) Gibberellin 3 beta-hydrox... 124 4e-29
TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-l... 120 8e-28
TC217747 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyg... 113 1e-25
TC220111 similar to PIR|T49209|T49209 leucoanthocyanidin dioxyge... 113 1e-25
TC215577 homologue to UP|Q41681 (Q41681) 1-aminocylopropane-1-ca... 110 9e-25
TC225031 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-c... 108 3e-24
>TC228680 similar to UP|G2O1_PEA (Q9SQ80) Gibberellin 2-beta-dioxygenase 1
(Gibberellin 2-beta-hydroxylase 1) (Gibberellin
2-oxidase 1) (GA 2-oxidase 1) (SLENDER protein) ,
complete
Length = 1226
Score = 288 bits (738), Expect = 2e-78
Identities = 155/310 (50%), Positives = 209/310 (67%), Gaps = 7/310 (2%)
Frame = +1
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+PIVDL K + LIVKA EE+GFFKVINHG+S E I+++E F FF+ + +K++
Sbjct: 241 IPIVDLS--KPDAKTLIVKACEEFGFFKVINHGVSMEAISELEYEAFKFFSMSLNEKEKV 414
Query: 79 AP----GYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYTEAVR 134
P GYG K IG NGD+G +EYLLLN N + +P FR +++Y +VR
Sbjct: 415 GPPNPFGYGSKKIGHNGDVGWIEYLLLNTNQEHNFSVYGK---NPEKFRCLLNSYMSSVR 585
Query: 135 ELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYP--PILNSNNREDKSPSYNN 192
++ACEILE+MAEGL +Q VFS+L+ D +SDS+ R+NHY P + N++ N
Sbjct: 586 KMACEILELMAEGLKIQQKDVFSKLLMDKQSDSIFRVNHYAACPEMTLNDQ--------N 741
Query: 193 KVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFV 252
+GFGEH+DPQI+++LRSN+ SGLQI L+DG WI V PD ++F +NVGD L+VMTNGRF
Sbjct: 742 LIGFGEHTDPQIISLLRSNNTSGLQIYLRDGNWISVPPDDKSFFINVGDSLQVMTNGRFR 921
Query: 253 SVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPV-LVTPERPSLFRPFTWADYKKATYSL 311
SVRHR + N FKSR+SM YFG PPL I PP+ + + SL++ FTW +YKK+ Y
Sbjct: 922 SVRHRVLANGFKSRLSMIYFGGPPLSEKI--PPLSSLMKGKESLYKEFTWFEYKKSIYGS 1095
Query: 312 RLGDSRMELF 321
RL +R+E F
Sbjct: 1096RLSKNRLEHF 1125
>TC207179 similar to UP|G2O1_PEA (Q9SQ80) Gibberellin 2-beta-dioxygenase 1
(Gibberellin 2-beta-hydroxylase 1) (Gibberellin
2-oxidase 1) (GA 2-oxidase 1) (SLENDER protein) ,
partial (80%)
Length = 1097
Score = 244 bits (623), Expect = 4e-65
Identities = 133/250 (53%), Positives = 174/250 (69%), Gaps = 7/250 (2%)
Frame = +3
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+PIVDL K + LIVKA EE+GFFKVINHG+ ETI+++E F FF+ P+ +K++A
Sbjct: 246 IPIVDLS--KPDAKTLIVKACEEFGFFKVINHGVPMETISQLESEAFKFFSMPLNEKEKA 419
Query: 79 AP----GYGCKNIGFNGDMGEVEYLLLNANTP-SIAQISKSVSIDPSNFRSTVSAYTEAV 133
P GYG K IG NGD+G VEYLLLN N + + K+ FR +++Y +V
Sbjct: 420 GPPKPYGYGSKKIGHNGDVGWVEYLLLNTNQEHNFSFYGKNAE----KFRCLLNSYMSSV 587
Query: 134 RELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYP--PILNSNNREDKSPSYN 191
R++ACEILE+MAEGL +Q VFS+L+ D +SDSVLR+NHYP P L N +
Sbjct: 588 RKMACEILELMAEGLKIQQKNVFSKLLMDKQSDSVLRVNHYPACPELAVNGQ-------- 743
Query: 192 NKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRF 251
N +GFGEH+DPQI+++LRSN+ SGLQI L+DG WI V PD ++F +NVGD L+VMTNGRF
Sbjct: 744 NLIGFGEHTDPQIISLLRSNNTSGLQIFLRDGNWISVPPDHKSFFINVGDSLQVMTNGRF 923
Query: 252 VSVRHRAMTN 261
SVRHR + N
Sbjct: 924 RSVRHRVLAN 953
>TC207180 similar to GB|AAD45425.1|5579094|AF100955 gibberellin 2-oxidase
{Pisum sativum;} , partial (61%)
Length = 867
Score = 212 bits (539), Expect = 2e-55
Identities = 108/200 (54%), Positives = 142/200 (71%)
Frame = +3
Query: 122 FRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSN 181
FR +++Y +VR++ACEILE+MAEGL +Q VFS+L+ D ESDSV R+NHYP
Sbjct: 18 FRCLLNSYMSSVRKMACEILELMAEGLKIQQKNVFSKLLMDKESDSVFRVNHYPACPELV 197
Query: 182 NREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGD 241
N + N +GFGEH+DPQI+++LRSN+ SGLQI L+DG WI V PD ++F +NVGD
Sbjct: 198 NGQ-------NMIGFGEHTDPQIISLLRSNNTSGLQIFLRDGNWISVPPDHKSFFINVGD 356
Query: 242 VLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTW 301
L+VMTNGRF SV+HR +TN FKSR+SM YFG PPL IV P + + SL++ FTW
Sbjct: 357 SLQVMTNGRFRSVKHRVLTNGFKSRLSMIYFGGPPLSEKIV-PLSSLMKGKESLYKEFTW 533
Query: 302 ADYKKATYSLRLGDSRMELF 321
+YK TY+ RL D+R+ F
Sbjct: 534 FEYKNLTYASRLADNRLGHF 593
>TC234107 similar to UP|G2O2_PEA (Q9XHM5) Gibberellin 2-beta-dioxygenase 2
(Gibberellin 2-beta-hydroxylase 2) (Gibberellin
2-oxidase 2) (GA 2-oxidase 2) , partial (40%)
Length = 687
Score = 187 bits (476), Expect = 4e-48
Identities = 86/132 (65%), Positives = 105/132 (79%)
Frame = +3
Query: 191 NNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGR 250
NN +G EHSDPQILTI+RSN+V GLQIS DG+WIPV PDP V VGD L+V+TNGR
Sbjct: 36 NNNIGXXEHSDPQILTIMRSNNVDGLQISTHDGLWIPVPPDPNEXXVMVGDALQVLTNGR 215
Query: 251 FVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYS 310
F SVRHR +TN+ K+RMSM YF APPL+ I P++VTP PSL++PFTWA YK+A YS
Sbjct: 216 FASVRHRVLTNTTKARMSMMYFAAPPLNRWITPLPMMVTPHNPSLYKPFTWAQYKQAAYS 395
Query: 311 LRLGDSRMELFR 322
LRLGD+R++LF+
Sbjct: 396 LRLGDARLDLFK 431
>BU549366 homologue to GP|4678586|emb GA 2-oxidase {Phaseolus coccineus},
partial (51%)
Length = 622
Score = 166 bits (419), Expect = 2e-41
Identities = 84/168 (50%), Positives = 110/168 (65%)
Frame = -2
Query: 157 SRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGL 216
SRL++D +SDS RLN PP E ++ + N VGFGEH+DPQI+++LRSN SGL
Sbjct: 606 SRLLKDEKSDSCFRLNXXPPC-----PEVQALNGRNLVGFGEHTDPQIISVLRSNSTSGL 442
Query: 217 QISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPP 276
QI L DG W+ V PD +F +NVGD L+VMTNGRF SV+HR + + KSR+SM YFG P
Sbjct: 441 QICLTDGTWVSVPPDQTSFFINVGDTLQVMTNGRFKSVKHRVLADPTKSRLSMIYFGGAP 262
Query: 277 LHACIVAPPVLVTPERPSLFRPFTWADYKKATYSLRLGDSRMELFRNN 324
L I P L+ S ++ FTW +YKKA Y+ RL D+R+ F +
Sbjct: 261 LSEKISPLPSLMLKGEESFYKEFTWWEYKKAAYASRLADNRLAPFEKS 118
>TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (52%)
Length = 1808
Score = 152 bits (383), Expect = 3e-37
Identities = 114/338 (33%), Positives = 168/338 (48%), Gaps = 12/338 (3%)
Frame = +1
Query: 2 VLASRNP-IRSEKIVPGDLPIVDLKAEKSEVTRLIVK---ASEEYGFFKVINHGISHETI 57
V A+++P I S I +PI+DL SE + K A +E+GFF++INHG+ +
Sbjct: 112 VHANQDPHILSNTISLPQVPIIDLHQLLSEDPSELEKLDHACKEWGFFQLINHGVDPPVV 291
Query: 58 AKMEEAGFGFFAKPMPQKK------QAAPGYGCKNIGFNGDMGEVEYLLLNANTPSIAQI 111
M+ FF PM +K+ + G+G + E + P ++
Sbjct: 292 ENMKIGVQEFFNLPMEEKQKFWQTPEDMQGFGQLFVVSEEQKLEWADMFYAHTFPLHSRN 471
Query: 112 SKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRL 171
+ P FR + Y +R++ I+ +M + L ++ T S L D +R+
Sbjct: 472 PHLIPKIPQPFRENLENYCLELRKMCITIIGLMKKALKIK-TNELSELFEDPSQG--IRM 642
Query: 172 NHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKP 230
N+YPP P +G HSD LTIL + N+V GLQI +DG WIPVKP
Sbjct: 643 NYYPPC----------PQPERVIGINPHSDSGALTILLQVNEVEGLQIR-KDGKWIPVKP 789
Query: 231 DPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTP 290
AF +NVGD+LE++TNG + S+ HR + NS K R+S+A F P + I P LVTP
Sbjct: 790 LSNAFVINVGDMLEILTNGIYRSIEHRGIVNSEKERISIAMFHRPQMSRVIGPAPSLVTP 969
Query: 291 ERPSLFRPFTWADYKKATYSLRL-GDSRMELFRNNNDM 327
ERP+LF+ ADY L G S M++ R N++
Sbjct: 970 ERPALFKRIGVADYLNGFLKRELKGKSYMDVIRIQNEI 1083
>TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (54%)
Length = 1070
Score = 149 bits (375), Expect = 2e-36
Identities = 101/309 (32%), Positives = 161/309 (51%), Gaps = 10/309 (3%)
Frame = +2
Query: 24 LKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKK------Q 77
+++ SE+ +L + A +E+GFF+++NHG++ + K+ FF PM +KK Q
Sbjct: 5 VESGSSELDKLHL-ACKEWGFFQLVNHGVNSSLVEKVRLETQDFFNLPMSEKKKFWQTPQ 181
Query: 78 AAPGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYTEAVRELA 137
G+G + + L P +++ P FR T+ AY+ +++LA
Sbjct: 182 HMEGFGQAFVVSEDQKLDWADLYYMTTLPKHSRMPHLFPQLPLPFRDTLEAYSREIKDLA 361
Query: 138 CEILEVMAEGLGVQDTLVFSRLIRDIESDSV--LRLNHYPPILNSNNREDKSPSYNNKVG 195
I+ +M + L +Q+ R IR++ D + +R+N+YPP P +G
Sbjct: 362 IVIIGLMGKALKIQE-----REIRELFEDGIQLMRMNYYPPC----------PEPEKVIG 496
Query: 196 FGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFVSV 254
HSD L IL + N+V GLQI +DG+W+PVKP AF VNVGD+LE++TNG + S+
Sbjct: 497 LTPHSDGIGLAILLQLNEVEGLQIR-KDGLWVPVKPLINAFIVNVGDILEIITNGIYRSI 673
Query: 255 RHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYSLRL- 313
HRA N K R+S A F +P + P L+T + P F+ DY K +S +L
Sbjct: 674 EHRATVNGEKERLSFATFYSPSSDGVVGPAPSLITEQTPPRFKSIGVKDYFKGLFSRKLD 853
Query: 314 GDSRMELFR 322
G + +E+ R
Sbjct: 854 GKAYIEVMR 880
>TC205377
Length = 1345
Score = 135 bits (339), Expect = 3e-32
Identities = 95/316 (30%), Positives = 152/316 (48%), Gaps = 30/316 (9%)
Frame = +3
Query: 19 LPIVDLK-------AEKSEVTRLIVK---ASEEYGFFKVINHGISHETIAKMEEAGFGFF 68
+PI+DL ++ S + L+ + A E+GFF+V NHG+ +E+A FF
Sbjct: 165 IPIIDLSPITNHRVSDPSAIEGLVKEIGSACNEWGFFQVTNHGVPLTLRQNIEKASKLFF 344
Query: 69 AKPMPQKKQAAPG-------YGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSID--- 118
A+ +K++ + Y ++ D EV L A P+ ++ D
Sbjct: 345 AQSAEEKRKVSRNESSPAGYYDTEHTKNVRDWKEVFDFL--AKEPTFIPVTSDEHDDRVN 518
Query: 119 ---------PSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVL 169
P NFR Y + + +L+ +ILE++A LG++ V I+D S +
Sbjct: 519 QWTNQSPEYPLNFRVVTQEYIQEMEKLSFKILELIALSLGLEAKRVEEFFIKD--QTSFI 692
Query: 170 RLNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQ-DGVWIPV 228
RLNHYPP P + +G G H DP LTIL ++V GL++ + D WI V
Sbjct: 693 RLNHYPPC----------PYPDLALGVGRHKDPGALTILAQDEVGGLEVRRKADQEWIRV 842
Query: 229 KPDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLV 288
KP P+A+ +N+GD ++V +N + SV HR + NS K R S+ +F P + L+
Sbjct: 843 KPTPDAYIINIGDTVQVWSNDAYESVDHRVVVNSEKERFSIPFFFFPAHDTEVKPLEELI 1022
Query: 289 TPERPSLFRPFTWADY 304
+ PS +RP+ W +
Sbjct: 1023NEQNPSKYRPYKWGKF 1070
>TC209692 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (36%)
Length = 1229
Score = 131 bits (330), Expect = 4e-31
Identities = 102/329 (31%), Positives = 162/329 (49%), Gaps = 11/329 (3%)
Frame = +2
Query: 9 IRSEKIVPGDLPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFF 68
I +K++ G+ D++ E ++T A ++GFF+++ HGIS + +E+ GFF
Sbjct: 146 INLKKLIHGE----DIELELEKLT----SACRDWGFFQLVEHGISSVVMKTLEDEVEGFF 301
Query: 69 AKPMPQKKQA------APGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNF 122
PM +K + GYG + + + L + N SI + PS+
Sbjct: 302 MLPMEEKMKYKVRPGDVEGYGTVIGSEDQKLDWGDRLFMKINXRSIRNPHLFPEL-PSSL 478
Query: 123 RSTVSAYTEAVRELACEILEVMAEGLGVQ--DTLVFSRLIRDIESDSVLRLNHYPPILNS 180
R+ + Y E ++ LA ++ ++ + L ++ + VF I++ +R+ +YPP
Sbjct: 479 RNILELYIEELQNLAMILMGLLGKTLKIEKRELEVFEDGIQN------MRMTYYPPC--- 631
Query: 181 NNREDKSPSYNNKVGFGEHSDPQILTILRS-NDVSGLQISLQDGVWIPVKPDPEAFCVNV 239
P +G HSD +TIL N V+GLQI +DGVWIPV EA VN+
Sbjct: 632 -------PQPELVMGLSAHSDATGITILNQMNGVNGLQIK-KDGVWIPVNVISEALVVNI 787
Query: 240 GDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPF 299
GD++E+M+NG + SV HRA NS K R+S+A F P + I L PE P LF+
Sbjct: 788 GDIIEIMSNGAYKSVEHRATVNSEKERISVAMFFLPKFQSEIGPAVSLTNPEHPPLFKRI 967
Query: 300 TWADYKK--ATYSLRLGDSRMELFRNNND 326
+Y K T++ G S +E R +D
Sbjct: 968 VVEEYIKDYFTHNKLNGKSYLEHMRITDD 1054
>TC226020 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(93%)
Length = 1441
Score = 128 bits (321), Expect = 4e-30
Identities = 88/307 (28%), Positives = 152/307 (48%), Gaps = 8/307 (2%)
Frame = +3
Query: 21 IVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQAAP 80
I ++ + E+ IV+A E +G F+V++HG+ + +A+M FFA P P +K
Sbjct: 204 IDEVDGRRREICEKIVEACENWGIFQVVDHGVDQQLVAEMTRLAKEFFALP-PDEKLRFD 380
Query: 81 GYGCKNIGF-------NGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYTEAV 133
G K GF + + ++ + P + P +RS Y++ V
Sbjct: 381 MSGAKKGGFIVSSHLQGESVQDWREIVTYFSYPKRERDYSRWPDTPEGWRSVTEEYSDKV 560
Query: 134 RELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYNNK 193
LAC+++EV++E +G++ + + + D + +N+YP K P +
Sbjct: 561 MGLACKLMEVLSEAMGLEKEGLSKACV---DMDQKVVVNYYP----------KCPQPDLT 701
Query: 194 VGFGEHSDPQILTILRSNDVSGLQISLQDG-VWIPVKPDPEAFCVNVGDVLEVMTNGRFV 252
+G H+DP +T+L + V GLQ + +G WI V+P AF VN+GD ++NGRF
Sbjct: 702 LGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEAAFVVNLGDHAHYLSNGRFK 881
Query: 253 SVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYSLR 312
+ H+A+ NS SR+S+A F P +A V P + E+P + P T+A+ + S
Sbjct: 882 NADHQAVVNSNHSRLSIATFQNPAPNA-TVYPLKIREGEKPVMEEPITFAEMYRRKMSKD 1058
Query: 313 LGDSRME 319
+ +RM+
Sbjct: 1059IEIARMK 1079
>TC220711 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (27%)
Length = 837
Score = 127 bits (318), Expect = 9e-30
Identities = 74/184 (40%), Positives = 104/184 (56%), Gaps = 1/184 (0%)
Frame = +1
Query: 122 FRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSN 181
+ + Y +R+LA + ++ + LG + + L ES +R+N+YPP
Sbjct: 88 YHNLSEVYCNEMRDLAINMYVLIGKALGTEPNEIKDTLG---ESGQAIRINYYPPC---- 246
Query: 182 NREDKSPSYNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPDPEAFCVNVG 240
P N +G H+D LTIL + N+V GLQI +DG W+PVKP P AF V++G
Sbjct: 247 ------PQPENVLGLNAHTDASALTILLQGNEVEGLQIK-KDGTWVPVKPLPNAFIVSLG 405
Query: 241 DVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFT 300
DVLEV+TNG + S HRA+ NS K R+S+A F P A I P +VTPERP+LF+
Sbjct: 406 DVLEVVTNGIYKSSEHRAVVNSQKERLSIATFSGPEWSANIGPTPSVVTPERPALFKTIG 585
Query: 301 WADY 304
AD+
Sbjct: 586 VADF 597
>TC228068 anthocyanidin synthase [Glycine max]
Length = 1231
Score = 126 bits (316), Expect = 2e-29
Identities = 94/314 (29%), Positives = 149/314 (46%), Gaps = 16/314 (5%)
Frame = +3
Query: 7 NPIRSEKIVPGDLPIVDLKA--EKSEVTR-----LIVKASEEYGFFKVINHGISHETIAK 59
N EK +P +DL+ + EV R + KA+EE+G ++NHGI E I +
Sbjct: 105 NVFEEEKKEGLQVPTIDLREIDSEDEVVRGKCREKLKKAAEEWGVMHLVNHGIPDELIER 284
Query: 60 MEEAGFGFFAKPMPQKKQAA--------PGYGCKNIGFNGDMGEVEYLLLNANTPSIAQI 111
+++AG FF + +K++ A GYG K E E + P +
Sbjct: 285 VKKAGETFFGLAVEEKEKYANDLESGKIQGYGSKLANNASGQLEWEDYFFHLAFPEDKRD 464
Query: 112 SKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRL 171
P+++ S Y + +R LA +ILE ++ GLG++ + + E L++
Sbjct: 465 LSFWPKKPADYIEVTSEYAKRLRGLATKILEALSIGLGLEGRRLEKEVGGMEELLLQLKI 644
Query: 172 NHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPD 231
N+YP P +G H+D LT L N V GLQ+ Q G W+ K
Sbjct: 645 NYYPIC----------PQPELALGVEAHTDVSSLTFLLHNMVPGLQLFYQ-GQWVTAKCV 791
Query: 232 PEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAP-PVLVTP 290
P + +++GD +E+++NG++ S+ HR + N K R+S A F PP I+ P P LVT
Sbjct: 792 PNSILMHIGDTIEILSNGKYKSILHRGLVNKEKVRISWAVFCEPPKEKIILQPLPELVTE 971
Query: 291 ERPSLFRPFTWADY 304
P+ F P T+A +
Sbjct: 972 TEPARFPPRTFAQH 1013
>TC231312 weakly similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (34%)
Length = 845
Score = 126 bits (316), Expect = 2e-29
Identities = 77/202 (38%), Positives = 111/202 (54%), Gaps = 2/202 (0%)
Frame = +1
Query: 123 RSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNN 182
R + Y+ +++L I+E MA L ++ + + D+ +R +YPP
Sbjct: 10 RDNIENYSSQLKKLCLTIIERMAMALKIESNELLDYVFEDVFQ--TMRWTYYPPC----- 168
Query: 183 REDKSPSYNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGD 241
P N +G HSD LTIL ++N+ GLQI +DG WIPVKP P AF +NVGD
Sbjct: 169 -----PQPENVIGINPHSDACALTILLQANETEGLQIK-KDGNWIPVKPLPNAFVINVGD 330
Query: 242 VLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTW 301
+LE++TNG + S+ HRA N K R+S+A F P ++ I P LVT ER ++F+
Sbjct: 331 ILEILTNGIYRSIEHRATINKEKERISVATFHRPLMNKVIGPTPSLVTSERAAVFKRIAV 510
Query: 302 ADYKKATYSLRL-GDSRMELFR 322
DY KA +S L G S ++L R
Sbjct: 511 EDYYKAYFSRGLKGKSCLDLIR 576
>TC216796 similar to GB|AAQ65160.1|34365697|BT010537 At4g10500 {Arabidopsis
thaliana;} , partial (72%)
Length = 1454
Score = 124 bits (312), Expect = 4e-29
Identities = 88/301 (29%), Positives = 154/301 (50%), Gaps = 15/301 (4%)
Frame = +1
Query: 19 LPIVDLK----AEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQ 74
+P++DL+ +S + + I +A + YGFF+V NHG+ I K+ + FF P +
Sbjct: 214 IPLIDLQDLHGPNRSHIIQQIDQACQNYGFFQVTNHGVPEGVIEKIMKVTREFFGLPESE 393
Query: 75 KKQAAPGYGCK----NIGFNGDMGEV----EYLLLNANTPSIAQISKSVSIDPSNFRSTV 126
K ++ K + FN + +V ++L L+ + I K +P + R V
Sbjct: 394 KLKSYSTDPFKASRLSTSFNVNSEKVSSWRDFLRLHCHP--IEDYIKEWPSNPPSLREDV 567
Query: 127 SAYTEAVRELACEILEVMAEGLGVQDTLVFSRLI--RDIESDSVLRLNHYPPILNSNNRE 184
+ Y +R ++ +++E ++E LG++ + +R++ + + L +N+YP
Sbjct: 568 AEYCRKMRGVSLKLVEAISESLGLERDYI-NRVVGGKKGQEQQHLAMNYYPAC------- 723
Query: 185 DKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLE 244
P G H+DP ++TIL ++V GLQ+ L+DG W+ V P P F VNVGD ++
Sbjct: 724 ---PEPELTYGLPGHTDPTVITILLQDEVPGLQV-LKDGKWVAVNPIPNTFVVNVGDQIQ 891
Query: 245 VMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACI-VAPPVLVTPERPSLFRPFTWAD 303
V++N ++ SV HRA+ N K R+S+ F P A I AP ++ P + FT+ +
Sbjct: 892 VISNDKYKSVLHRAVVNCNKDRISIPTFYFPSNDAIIGPAPQLIHHHHHPPQYNNFTYNE 1071
Query: 304 Y 304
Y
Sbjct: 1072Y 1074
>TC230772 similar to UP|O24648 (O24648) Gibberellin 3 beta-hydroxylase
(2-oxoglutarate-dependent dioxygenase), partial (85%)
Length = 1264
Score = 124 bits (312), Expect = 4e-29
Identities = 85/296 (28%), Positives = 145/296 (48%), Gaps = 10/296 (3%)
Frame = +3
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+P++DL + ++LI A +G ++V+NHGI + ++ G F+ P QK +A
Sbjct: 99 VPVIDLNDPNA--SKLIHHACTTWGAYQVLNHGIPMSLLQDIQWVGETLFSLPSHQKHKA 272
Query: 79 A------PGYGCKNIG--FNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYT 130
A GYG I F M + ++ + Q+ D + V Y
Sbjct: 273 ARSPDGVDGYGLARISSFFPKLMWSEGFTIVGSPLEHFRQLWPQ---DYDKYCDFVMQYD 443
Query: 131 EAVRELACEILEVMAEGLGV-QDTLVFSRLIRDIESD-SVLRLNHYPPILNSNNREDKSP 188
EA+++L +++ +M + LG+ ++ L ++ E + L+LN YP P
Sbjct: 444 EAMKKLVGKLMLLMLDSLGITKEDLKWAGSKGQFEKTCAALQLNSYPTC----------P 593
Query: 189 SYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTN 248
+ +G H+D +LTIL N++SGLQ+ + W+ V P +NVGD+L +++N
Sbjct: 594 DPDRAMGLAAHTDSTLLTILYQNNISGLQVHRKGVGWVTVPPLSGGLVINVGDLLHILSN 773
Query: 249 GRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADY 304
G + SV HR + N + R+S+AY PP + I LV P +P L++ TW +Y
Sbjct: 774 GLYPSVLHRVLVNRIQRRLSVAYLCGPPPNVEICPHAKLVGPNKPPLYKAVTWNEY 941
>TC205989 similar to UP|Q9FLV0 (Q9FLV0) Flavanone 3-hydroxylase-like protein,
partial (74%)
Length = 1192
Score = 120 bits (301), Expect = 8e-28
Identities = 89/275 (32%), Positives = 136/275 (49%), Gaps = 10/275 (3%)
Frame = +2
Query: 59 KMEEAGFGFFAKPMPQKKQAAPGYGCKNI----GFNGDMGEV----EYLLLNANTPSIAQ 110
+MEE GFF P+ +K + K + FN V +YL L+ + +
Sbjct: 8 EMEEVAHGFFKLPVEEKLKLYSEDTSKTMRLSTSFNVKKETVRNWRDYLRLHCYP--LEK 181
Query: 111 ISKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLR 170
+ +P +F+ TV+ Y +REL I E ++E LG++ + + L E +
Sbjct: 182 YAPEWPSNPPSFKETVTEYCTIIRELGLRIQEYISESLGLEKDYIKNVLG---EQGQHMA 352
Query: 171 LNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSN-DVSGLQISLQDGVWIPVK 229
+N+YPP P G H+DP LTIL + V+GLQ+ L+DG W+ V
Sbjct: 353 VNYYPPC----------PEPELTYGLPGHTDPNALTILLQDLQVAGLQV-LKDGKWLAVS 499
Query: 230 PDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVT 289
P P AF +N+GD L+ ++NG + SV HRA+ N K R+S+A F P A I L
Sbjct: 500 PQPNAFVINIGDQLQALSNGLYKSVWHRAVVNVEKPRLSVASFLCPNDEALISPAKPLTE 679
Query: 290 PERPSLFRPFTWADYKKATYSLRLGDSR-MELFRN 323
+++R FT+A+Y K +S L +ELF+N
Sbjct: 680 HGSEAVYRGFTYAEYYKKFWSRNLDQEHCLELFKN 784
>TC217747 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (53%)
Length = 713
Score = 113 bits (283), Expect = 1e-25
Identities = 68/188 (36%), Positives = 102/188 (54%), Gaps = 1/188 (0%)
Frame = +1
Query: 119 PSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPIL 178
P R ++ Y E V + IL++M+ LG+++ + + + E + LR+N YP
Sbjct: 16 PEXXRKVIAEYGEGVVKXGGRILKMMSINLGLKEDFLLNAFGGESEVGACLRVNFYP--- 186
Query: 179 NSNNREDKSPSYNNKVGFGEHSDPQILTILRSND-VSGLQISLQDGVWIPVKPDPEAFCV 237
K P + G HSDP +TIL +D VSGLQ+ D WI VKP P AF +
Sbjct: 187 -------KCPQPDLTFGLSPHSDPGGMTILLPDDFVSGLQVRRGDE-WITVKPVPNAFII 342
Query: 238 NVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFR 297
N+GD ++V++N + SV HR + NS K R+S+A F P I LVT E+P+L+
Sbjct: 343 NIGDQIQVLSNAIYKSVEHRVIVNSNKDRVSLALFYNPRSDLLIQPAKELVTEEKPALYS 522
Query: 298 PFTWADYK 305
P T+ +Y+
Sbjct: 523 PMTYDEYR 546
>TC220111 similar to PIR|T49209|T49209 leucoanthocyanidin dioxygenase-like
protein - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (43%)
Length = 786
Score = 113 bits (282), Expect = 1e-25
Identities = 66/188 (35%), Positives = 107/188 (56%), Gaps = 1/188 (0%)
Frame = +3
Query: 119 PSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPIL 178
P++ R+ +S Y E V L ILE+++ LG+++ + + + + + LR+N YP
Sbjct: 15 PTSLRNIISEYGEQVVMLGGRILEILSINLGLREDFLLNAFGGENDLGACLRVNFYP--- 185
Query: 179 NSNNREDKSPSYNNKVGFGEHSDPQILTILRSND-VSGLQISLQDGVWIPVKPDPEAFCV 237
K P + +G HSDP +TIL ++ VSGLQ+ + WI VKP P AF +
Sbjct: 186 -------KCPQPDLTLGLSPHSDPGGMTILLPDENVSGLQVRRGED-WITVKPVPNAFII 341
Query: 238 NVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFR 297
N+GD ++V++N + S+ HR + NS K R+S+A+F P I LVT +RP+L+
Sbjct: 342 NMGDQIQVLSNAIYKSIEHRVIVNSDKDRVSLAFFYNPRSDIPIQPAKELVTKDRPALYP 521
Query: 298 PFTWADYK 305
P T+ +Y+
Sbjct: 522 PMTFDEYR 545
>TC215577 homologue to UP|Q41681 (Q41681) 1-aminocylopropane-1-carboxylate
oxidase homolog , complete
Length = 1382
Score = 110 bits (275), Expect = 9e-25
Identities = 86/317 (27%), Positives = 156/317 (49%), Gaps = 10/317 (3%)
Frame = +2
Query: 18 DLPIVDL----KAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMP 73
+ P+VD+ E+ +I A E +GFF+++NHGIS E + +E+ + K M
Sbjct: 56 NFPVVDMGKLNTEERPAAMEIIKDACENWGFFELVNHGISIELMDTVEKLTKEHYKKTME 235
Query: 74 QK-KQAAPGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYTEA 132
Q+ K+ G +++ + + E + P ++ +S + +D ++R T+ +
Sbjct: 236 QRFKEMVTSKGLESVQSEINDLDWESTFFLRHLP-LSNVSDNADLD-QDYRKTMKKFALE 409
Query: 133 VRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYNN 192
+ +LA ++L+++ E LG++ + + ++++YPP P+ +
Sbjct: 410 LEKLAEQLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPC----------PTPDL 559
Query: 193 KVGFGEHSDPQ-ILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRF 251
G H+D I+ + + + VSGLQ+ L+D WI V P + +N+GD LEV+TNG++
Sbjct: 560 IKGLRAHTDAGGIILLFQDDKVSGLQL-LKDDQWIDVPPMRHSIVINLGDQLEVITNGKY 736
Query: 252 VSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLV--TPERPSLFRPFTWADYKK--A 307
SV HR + + +RMS+A F P A I P LV E ++ F + DY K A
Sbjct: 737 KSVMHRVIAQADDTRMSIASFYNPGDDAVISPAPALVKELDETSQVYPKFVFDDYMKLYA 916
Query: 308 TYSLRLGDSRMELFRNN 324
+ + R E + N
Sbjct: 917 GLKFQAKEPRFEAMKAN 967
>TC225031 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1244
Score = 108 bits (270), Expect = 3e-24
Identities = 84/321 (26%), Positives = 152/321 (47%), Gaps = 13/321 (4%)
Frame = +1
Query: 18 DLPIVDLKA----EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMP 73
+ P+++L+ E+++ I A E +GFF+++NHGI H+ + +E + K M
Sbjct: 118 NFPLINLEKLSGEERNDTMEKIKDACENWGFFELVNHGIPHDILDTVERLTKEHYRKCME 297
Query: 74 QKKQ---AAPGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYT 130
++ + A+ G E + L + +I++I + +R + +
Sbjct: 298 ERFKELVASKGLDAVQTEVKDMDWESTFHLRHLPESNISEIPDLID----EYRKVMKDFA 465
Query: 131 EAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSY 190
+ +LA ++L+++ E LG++ + ++ +YPP P+
Sbjct: 466 LRLEKLAEQLLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPPC----------PNP 615
Query: 191 NNKVGFGEHSDPQ-ILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNG 249
G H+D I+ + + + VSGLQ+ L+DG W+ V P + VN+GD LEV+TNG
Sbjct: 616 ELVKGLRPHTDAGGIILLFQDDKVSGLQL-LKDGQWVDVPPMRHSIVVNIGDQLEVITNG 792
Query: 250 RFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLV---TPERPSLFRPFTWADYKK 306
++ SV HR + + +RMS+A F P A I P L+ E+ L+ F + DY K
Sbjct: 793 KYKSVEHRVIAQTDGTRMSIASFYNPGSDAVIYPAPELLEKEAEEKNQLYPKFVFEDYMK 972
Query: 307 --ATYSLRLGDSRMELFRNNN 325
A + + R E F+ +N
Sbjct: 973 LYAKLKFQAKEPRFEAFKASN 1035
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,824,572
Number of Sequences: 63676
Number of extensions: 168734
Number of successful extensions: 1012
Number of sequences better than 10.0: 201
Number of HSP's better than 10.0 without gapping: 919
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 922
length of query: 327
length of database: 12,639,632
effective HSP length: 98
effective length of query: 229
effective length of database: 6,399,384
effective search space: 1465458936
effective search space used: 1465458936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0333b.12


