

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0331c.2
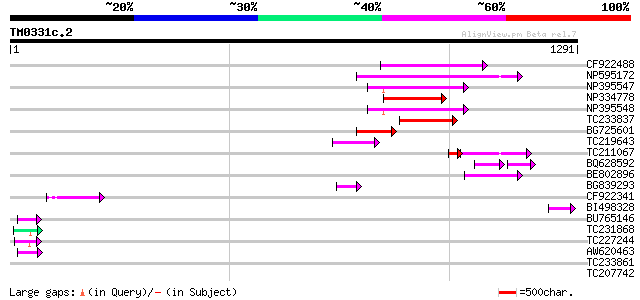
(1291 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922488 201 2e-51
NP595172 polyprotein [Glycine max] 187 2e-47
NP395547 reverse transcriptase [Glycine max] 158 2e-38
NP334778 reverse transcriptase [Glycine max] 152 1e-36
NP395548 reverse transcriptase [Glycine max] 145 9e-35
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 135 1e-31
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 100 5e-21
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 80 4e-15
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 58 3e-12
BQ628592 50 4e-12
BE802896 65 3e-10
BG839293 55 3e-07
CF922341 50 8e-06
BI498328 47 5e-05
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 46 9e-05
TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein... 44 4e-04
TC227244 similar to UP|Q941A9 (Q941A9) At1g26300/F28B23_4, parti... 44 6e-04
AW620463 44 6e-04
TC233861 weakly similar to UP|Q41192 (Q41192) NaPRP3, partial (21%) 43 0.001
TC207742 similar to UP|Q9FYG0 (Q9FYG0) F1N21.15 (At1g67330), par... 42 0.001
>CF922488
Length = 741
Score = 201 bits (511), Expect = 2e-51
Identities = 108/245 (44%), Positives = 147/245 (59%)
Frame = +3
Query: 844 VKKANGKWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHP 903
V K +GK MCVDY DLN A PKD +PLP I+ LVD + S MD +SG +QI++ P
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 904 ADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVR 963
D +KT F+T +CY+ M FGLKN GATYQR M +F + + +EVY+DDMIVKS
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 964 GLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMK 1023
+H +L + F +RK+ +RLNP KC F V+ K L F+ + RGIE++ K K I +M
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 1024 SPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLS 1083
P K+VQ GR+ + RF+ P F L KN +W +C AF R+K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 1084 SPPIL 1088
+P +L
Sbjct: 723 NPHVL 737
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 187 bits (476), Expect = 2e-47
Identities = 116/379 (30%), Positives = 187/379 (48%)
Frame = +1
Query: 790 HRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG 849
H + L PV R + +++ + ++L I+ P L +++VKK +G
Sbjct: 1759 HAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLP-ILLVKKKDG 1935
Query: 850 KWRMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKT 909
WR C DY LN KDS+P+P++D L+D G + S +D SG HQI + P D +KT
Sbjct: 1936 SWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKT 2115
Query: 910 AFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQ 969
AF T +Y + MPFGL NA AT+Q LM+++F + + + V+ DD+++ S DH +
Sbjct: 2116 AFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLK 2295
Query: 970 DLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVK 1029
LE +++H + KC FG +LG ++ G+ + K +A+ +P+NVK
Sbjct: 2296 HLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVK 2475
Query: 1030 EVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILS 1089
+++ G RF+ + + P L+K+ +F W E E AFV+LK+ ++ P+LS
Sbjct: 2476 QLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKD-SFLWNNEAEAAFVKLKKAMTEAPVLS 2652
Query: 1090 KPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLV 1149
P P L S + +V+ Q H I YF S L + + LA+
Sbjct: 2653 LPDFSQPFILETDASGIGVGAVLGQ---NGHPIAYF-SKKLAPRMQKQSAYTRELLAITE 2820
Query: 1150 TARRLRPYFQSFPVKVRTD 1168
+ R Y +RTD
Sbjct: 2821 ALSKFRHYLLGNKFIIRTD 2877
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 158 bits (399), Expect = 2e-38
Identities = 90/248 (36%), Positives = 129/248 (51%), Gaps = 18/248 (7%)
Frame = +1
Query: 816 VQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG------------------KWRMCVDY 857
V++EV KLL A I + +W++ V +V K G +WRMC+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 858 TDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARAN 917
LN+A KD YPLP +D ++ + +D YSG +QI + P D++KTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 918 YCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGE 977
+ YR MPFGL NA T+QR M +F V + +EV++DD + +LE+
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 978 IRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGR 1037
K N+ LN EKC+F VQ G LG I+ RGIE+ EK I ++ P NVK + G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 1038 IAALSRFL 1045
+ RF+
Sbjct: 721 VGFYRRFI 744
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 152 bits (383), Expect = 1e-36
Identities = 71/143 (49%), Positives = 96/143 (66%)
Frame = +3
Query: 852 RMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAF 911
RMCVDY DLN+A PKD++PLP ID L+ + L S MD +SG +QI+M P D +KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 912 MTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 971
+T +CY+ M FGLKN GATY R M +F + + +E YVD+MI KS +H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 972 EEAFGEIRKHNMRLNPEKCYFGV 994
+ FG++RK+ +RLNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 145 bits (367), Expect = 9e-35
Identities = 84/248 (33%), Positives = 132/248 (52%), Gaps = 18/248 (7%)
Frame = +1
Query: 816 VQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG------------------KWRMCVDY 857
V++EV KLL I + W++ V++V K G W++C+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 858 TDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGNHQIRMHPADEDKTAFMTARAN 917
LN+A KD +PLP +D +++ +G+ +DAY G +QI + P D++K AF
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 918 YCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGE 977
+ YR +PFGL NA T+Q M +FA V +++EV++DD V + LE
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 978 IRKHNMRLNPEKCYFGVQGGKFLGFMITSRGIEINPEKCKAIQQMKSPSNVKEVQRLTGR 1037
+ N+ LN EKC+F V+ G LG I++RGIE++ K I+++ PSNVK ++ G+
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 1038 IAALSRFL 1045
RF+
Sbjct: 721 ARFYRRFI 744
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 135 bits (340), Expect = 1e-31
Identities = 66/132 (50%), Positives = 90/132 (68%)
Frame = +2
Query: 888 SLMDAYSGNHQIRMHPADEDKTAFMTARANYCYRTMPFGLKNAGATYQRLMDRVFAGQVG 947
S MD +SG +QI M D +KT F+T + YR M FGLKN GATYQR M +F +
Sbjct: 5 SFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMMH 184
Query: 948 RNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCYFGVQGGKFLGFMITSR 1007
+ +EVYVDDMI KS +H +L + FG ++K+ ++LNP KC FGV+ GK LGF+++ +
Sbjct: 185 KEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQK 364
Query: 1008 GIEINPEKCKAI 1019
GIEI+PEK KA+
Sbjct: 365 GIEIDPEKVKAL 400
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 100 bits (248), Expect = 5e-21
Identities = 48/91 (52%), Positives = 66/91 (71%)
Frame = -3
Query: 790 HRLALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVIVKKANG 849
H+LA+ +K V+ +R++ ++ + V QEV KL A FIR++ Y T L +VV+VKK NG
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 850 KWRMCVDYTDLNKACPKDSYPLPSIDSLVDG 880
KWR+C DY DLN ACPKD+YPLP+ID + DG
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTDG 5
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 80.5 bits (197), Expect = 4e-15
Identities = 43/111 (38%), Positives = 62/111 (55%), Gaps = 4/111 (3%)
Frame = +3
Query: 736 EETKALKFGD----RTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHR 791
EET+ + G R +KIGT +T L LL + D+FAWS +DMPG+ + + HR
Sbjct: 978 EETELVDLGSGSGKREVKIGTGITAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHR 1157
Query: 792 LALNPSLKPVS*LRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVV 842
L LNP PV RR+ + +++EV K A F+ +YP W+AN+V
Sbjct: 1158 LPLNPECSPVKQKLRRMKPETSLKIKEEVKK*FDAGFLAVARYPKWVANIV 1310
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 57.8 bits (138), Expect(2) = 3e-12
Identities = 43/162 (26%), Positives = 73/162 (44%), Gaps = 1/162 (0%)
Frame = +1
Query: 1027 NVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPP 1086
+V +++ G + RF+P + P + ++KN+AF W + E+AF LKE L+ P
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAP 288
Query: 1087 ILSKPIQGHPLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALA 1146
+L+ P L S + +V+LQ G H I YF S L A + Y +K A
Sbjct: 289 VLALPDFSKTFELECDASGVGVRAVLLQ---GGHPIAYF-SEKLHSATLNYPTYDKELYA 456
Query: 1147 VLVTARRLRPYFQSFPVKVRTD-LPLRQVLQKPDLSGRLVAW 1187
++ + + + +D L+ + K L+ R W
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKW 582
Score = 33.5 bits (75), Expect(2) = 3e-12
Identities = 14/33 (42%), Positives = 22/33 (66%)
Frame = +2
Query: 999 FLGFMITSRGIEINPEKCKAIQQMKSPSNVKEV 1031
F GF++ G++++PEK KAIQ+ P V E+
Sbjct: 23 FSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI 121
>BQ628592
Length = 423
Score = 50.4 bits (119), Expect(2) = 4e-12
Identities = 23/71 (32%), Positives = 42/71 (58%), Gaps = 2/71 (2%)
Frame = -1
Query: 1059 LRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGHPLHLYFAVSDGALSSVMLQEIDG 1118
L KN A W + +EAF ++K+ L++P +L P+ G P LY + D ++ V++Q D
Sbjct: 423 LPKNQAILWNSNYQEAFEKIKQSLANPSVLMPPVTGRPFLLYMTMLDESMGCVLVQHDDS 244
Query: 1119 --EHRIVYFVS 1127
+ + +Y++S
Sbjct: 243 GKKEQAIYYLS 211
Score = 40.4 bits (93), Expect(2) = 4e-12
Identities = 18/64 (28%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Frame = -3
Query: 1134 EVRYQKIEKAALAVLVTARRLRPYFQSFPVKVRTDL-PLRQVLQKPDLSGRLVAWSVELS 1192
++ Y +E+ ++ + RLR Y S + + + P++ + +KP L+GR+ W V LS
Sbjct: 193 KMNYSMLERTCCTLVWASHRLRQYMLSHTTWLISKMDPVKYIFEKPALTGRIARWQVLLS 14
Query: 1193 EYSL 1196
E+++
Sbjct: 13 EFNI 2
>BE802896
Length = 416
Score = 64.7 bits (156), Expect = 3e-10
Identities = 40/133 (30%), Positives = 60/133 (45%)
Frame = -2
Query: 1036 GRIAALSRFLPKSGDRSFPFFKCLRKNVAFEWTAECEEAFVRLKELLSSPPILSKPIQGH 1095
G RF+ + P L+K V F++ +C+ AF LK L + PI+ P
Sbjct: 406 GHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPDWTA 227
Query: 1096 PLHLYFAVSDGALSSVMLQEIDGEHRIVYFVSHTLQGAEVRYQKIEKAALAVLVTARRLR 1155
P L S+ AL V+ Q+ID R++Y+ S TL A+ Y EK LA++ +
Sbjct: 226 PFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALEKFH 47
Query: 1156 PYFQSFPVKVRTD 1168
Y + V D
Sbjct: 46 SYLLGTRIIVYID 8
>BG839293
Length = 781
Score = 54.7 bits (130), Expect = 3e-07
Identities = 26/58 (44%), Positives = 35/58 (59%)
Frame = +1
Query: 744 GDRTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALNPSLKPV 801
G R +KIGT +T L LL + D+FAWS +DMPG+ + + H+L LNP PV
Sbjct: 550 GKREVKIGTGITAPIREELIILLKDYQDIFAWSYQDMPGLSSDIVQHQLPLNPECSPV 723
>CF922341
Length = 675
Score = 49.7 bits (117), Expect = 8e-06
Identities = 31/137 (22%), Positives = 62/137 (44%), Gaps = 4/137 (2%)
Frame = +1
Query: 84 HLMRSIGNIQQRNEHLQAQLDFYRREQRDDGSREADSVAEFRPFSED----VESVAIPDN 139
HL+ S + +N H+ A++ + + ++G R + ++ + + V ++ P
Sbjct: 250 HLLHSTTS---KNPHVMAEMG--KLDHLEEGLRAIEGGEDYAFANLEELFLVPNIITPPK 414
Query: 140 MKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSI 199
K L D Y G + PK+HL + KM A + + F + A+ W+T L +
Sbjct: 415 FKVLDFDKYKGTTCPKNHLKMYCQKMGAYAKDEELLIHSFQESLTGVAVTWYTNLEPSRV 594
Query: 200 SNFRDFSSKFLVQFSAN 216
+++D F+ Q+ N
Sbjct: 595 HSWKDLMVAFVRQYQYN 645
>BI498328
Length = 335
Score = 47.0 bits (110), Expect = 5e-05
Identities = 21/62 (33%), Positives = 34/62 (53%)
Frame = +1
Query: 1227 QWTLFVDGSSNSSGSGAGVTLEGPGELVLEQSLKFEFKATNNQAEYEALIAGLKLAREVK 1286
+W + DG+SN+ G G G L P + + + + F TNN AEYEA G++ A +
Sbjct: 148 KWIVCFDGASNALGHGVGAVLVSPDDQCIPFTARLGFDCTNNMAEYEACALGVQAAIDFD 327
Query: 1287 IR 1288
++
Sbjct: 328 VK 333
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 46.2 bits (108), Expect = 9e-05
Identities = 22/53 (41%), Positives = 24/53 (44%)
Frame = -1
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
PP P + L P +T P PPP PP PPPPPSP P P P
Sbjct: 260 PPLSPPSLXLSPP*KTPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPP 102
Score = 42.0 bits (97), Expect = 0.002
Identities = 20/57 (35%), Positives = 25/57 (43%)
Frame = -3
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P + P + +++P PPP PP PPPPP P P P P P
Sbjct: 276 PPPPPPPPLPPLPXSLSPLKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPPPP 106
Score = 40.8 bits (94), Expect = 0.004
Identities = 22/57 (38%), Positives = 23/57 (39%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P SP+ P PPP PP PPPPP PS P P P P
Sbjct: 361 PPPPPPPPPPPSPLPPPPPPXPPPPPPPP-PPPPPPPSPPPPXXSLPPKKPPPPPPP 194
Score = 40.8 bits (94), Expect = 0.004
Identities = 22/59 (37%), Positives = 24/59 (40%), Gaps = 2/59 (3%)
Frame = -3
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPP--PPSPSQVGSLERSPENSPAPEQQP 75
PP P SP P PPP PP PPP PP P + L+ P P P P
Sbjct: 354 PPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPLPPLPXSLSPLKNPPPPPPPPPPPP 178
Score = 40.8 bits (94), Expect = 0.004
Identities = 19/48 (39%), Positives = 21/48 (43%)
Frame = -1
Query: 28 LDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
L P + + SP PPP PP PPPPP P P P P P
Sbjct: 407 LPPPPPPSSLPSPPPPPPPPPPPPPPLPPPPPPPPXPPPPPPPPPPPP 264
Score = 40.4 bits (93), Expect = 0.005
Identities = 21/57 (36%), Positives = 22/57 (37%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P P+ P PPP PP PPPPPSP P P P P
Sbjct: 196 PPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPP 26
Score = 40.4 bits (93), Expect = 0.005
Identities = 21/57 (36%), Positives = 23/57 (39%)
Frame = -3
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P SP++ P PPP PP PPPPP P P P P P
Sbjct: 261 PPPLPPLPXSLSPLKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPPPPPLPPP 91
Score = 40.4 bits (93), Expect = 0.005
Identities = 20/58 (34%), Positives = 22/58 (37%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 76
PP P P + P PPP PP PPPPP P P P P P+
Sbjct: 262 PPPSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPS 89
Score = 40.4 bits (93), Expect = 0.005
Identities = 20/57 (35%), Positives = 21/57 (36%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P P + PPP PP PPPPP P L P P P P
Sbjct: 283 PPPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPP 113
Score = 40.4 bits (93), Expect = 0.005
Identities = 21/57 (36%), Positives = 22/57 (37%)
Frame = -3
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P P P PPP PP PPPPP P SP +P P P
Sbjct: 363 PPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPLPPLPXSLSPLKNPPPPPPP 193
Score = 40.4 bits (93), Expect = 0.005
Identities = 19/37 (51%), Positives = 19/37 (51%)
Frame = -2
Query: 39 SPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
SP PPP PP PPPPP PS L P P P P
Sbjct: 379 SPPPPPPPPPPPPPPPPS---PLPPPPPPXPPPPPPP 278
Score = 40.0 bits (92), Expect = 0.007
Identities = 23/65 (35%), Positives = 24/65 (36%)
Frame = -1
Query: 11 SPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPA 70
SP + PP P P P PPP PP PPPPPSP P P
Sbjct: 230 SPP*KTPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPSPPLPPPPPPPPPPPPP 51
Query: 71 PEQQP 75
P P
Sbjct: 50 PPPPP 36
Score = 40.0 bits (92), Expect = 0.007
Identities = 17/34 (50%), Positives = 18/34 (52%)
Frame = -1
Query: 40 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQ 73
P PPPSPP+PPPPP P P P P Q
Sbjct: 116 PPPPPSPPLPPPPPPPPPPPPPPPPPPPPPPPGQ 15
Score = 38.5 bits (88), Expect = 0.019
Identities = 17/39 (43%), Positives = 18/39 (45%)
Frame = -1
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQ 57
PP P P+ P PPP PP PPPPP P Q
Sbjct: 131 PPPPPPPPPPSPPLPPPPPPPPPPPPPPPPPPPPPPPGQ 15
Score = 37.7 bits (86), Expect = 0.033
Identities = 22/62 (35%), Positives = 25/62 (39%)
Frame = -1
Query: 17 ISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPA 76
+ PP P L SP P PPP PP PPPPP P P P P P
Sbjct: 407 LPPPPPPSS--LPSPPPPPPPPPPPPPPLPPPPPPPPXPP-------PPPPPPPPPPPPP 255
Query: 77 VT 78
++
Sbjct: 254 LS 249
Score = 37.4 bits (85), Expect = 0.043
Identities = 18/47 (38%), Positives = 21/47 (44%)
Frame = -1
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSP 65
PP P P + + P PPP PP PPPPP P + SP
Sbjct: 146 PPPPPPPPPPPPPPPSPPLPPPPPPPPPPPPPPPPPPPPPPPGQTSP 6
Score = 37.0 bits (84), Expect = 0.056
Identities = 19/53 (35%), Positives = 20/53 (36%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
PP P + P P PPP PP PPPPP P P P P
Sbjct: 178 PPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPP 20
Score = 36.2 bits (82), Expect = 0.096
Identities = 22/64 (34%), Positives = 27/64 (41%)
Frame = -2
Query: 12 PMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
P+ SPP P P + + P PP PP PPPPP P S P S P
Sbjct: 397 PLLPPPSPPPPPPPPPPPPPPPPSPLPPPPPPXPPPPPPPPPPPPPPPS-PPPPXXSLPP 221
Query: 72 EQQP 75
++ P
Sbjct: 220 KKPP 209
Score = 35.8 bits (81), Expect = 0.13
Identities = 19/53 (35%), Positives = 19/53 (35%)
Frame = -3
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
PP P P P PPP PP PPPPP P P P P
Sbjct: 180 PPPPPPPPFPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPPPPPP 22
Score = 35.8 bits (81), Expect = 0.13
Identities = 19/57 (33%), Positives = 21/57 (36%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P + P P PPP PP PPPP P P P P +P
Sbjct: 181 PPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPPGKP 11
Score = 35.8 bits (81), Expect = 0.13
Identities = 18/57 (31%), Positives = 21/57 (36%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P + P P PPP PP P PPP + + P P P P
Sbjct: 346 PPPPPPSPLPPPPPPXPPPPPPPPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPPPP 176
Score = 35.4 bits (80), Expect = 0.16
Identities = 19/57 (33%), Positives = 20/57 (34%)
Frame = -2
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
PP P + P P PPP PP PPPP S P P P P
Sbjct: 343 PPPPPSPLPPPPPPXPPPPPPPPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPPPPP 173
Score = 33.9 bits (76), Expect = 0.48
Identities = 14/32 (43%), Positives = 14/32 (43%)
Frame = -2
Query: 40 PSPPPSPPVPPPPPSPSQVGSLERSPENSPAP 71
P P P PP PPPPP P P P P
Sbjct: 100 PPPSPPPPPPPPPPPPPPPPPPPPPPPGKPLP 5
Score = 30.8 bits (68), Expect = 4.0
Identities = 21/73 (28%), Positives = 22/73 (29%), Gaps = 16/73 (21%)
Frame = -1
Query: 19 PPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPP----------------PPPSPSQVGSLE 62
PP P L P P PPP PP PP PPP P
Sbjct: 356 PPPPPPPPPLPPPPPPPPXPPPPPPPPPPPPPPPLSPPSLXLSPP*KTPPPPPPPPPPPP 177
Query: 63 RSPENSPAPEQQP 75
P P+P P
Sbjct: 176 PPPPPPPSPPPPP 138
>TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein
(Corresponding sequence E02A10.2), partial (23%)
Length = 790
Score = 44.3 bits (103), Expect = 4e-04
Identities = 27/83 (32%), Positives = 33/83 (39%), Gaps = 16/83 (19%)
Frame = -3
Query: 9 HHSPMRQRISPPRQPHRVVLDSPVRTTGVQSPSPPPSP----------------PVPPPP 52
H +R PP QP + +L + Q P PPP P P P PP
Sbjct: 482 HLLSIRSPPQPPPQPPQPLLSNLFPHPPPQPPQPPPQPPPQPAPQPPLLIKFPQPPPQPP 303
Query: 53 PSPSQVGSLERSPENSPAPEQQP 75
P P Q L + P+ P PE QP
Sbjct: 302 PQPPQPPPLIKFPQPPPQPEPQP 234
Score = 38.1 bits (87), Expect = 0.025
Identities = 25/79 (31%), Positives = 32/79 (39%), Gaps = 13/79 (16%)
Frame = -3
Query: 10 HSPMRQRISPPRQPHRVVLDSPVRTTGVQSP-SPPPSPPVPPP-------PPSPSQVGSL 61
H P + PP+ P + P+ Q P PPP PP PPP PP P L
Sbjct: 407 HPPPQPPQPPPQPPPQPAPQPPLLIKFPQPPPQPPPQPPQPPPLIKFPQPPPQPEPQPPL 228
Query: 62 -----ERSPENSPAPEQQP 75
+ P+ +P P Q P
Sbjct: 227 LIIFPQPPPQPAPQPPQPP 171
Score = 30.4 bits (67), Expect = 5.3
Identities = 16/53 (30%), Positives = 20/53 (37%), Gaps = 1/53 (1%)
Frame = -3
Query: 24 HRVVLDSPVRTTGVQSPSPPPSPPVPPP-PPSPSQVGSLERSPENSPAPEQQP 75
H S ++ + + SPP PPP PP P P P P QP
Sbjct: 527 HETYFSSSLKCSSIPHLLSIRSPPQPPPQPPQPLLSNLFPHPPPQPPQPPPQP 369
>TC227244 similar to UP|Q941A9 (Q941A9) At1g26300/F28B23_4, partial (67%)
Length = 1408
Score = 43.5 bits (101), Expect = 6e-04
Identities = 26/75 (34%), Positives = 34/75 (44%), Gaps = 15/75 (20%)
Frame = +1
Query: 12 PMRQRISPPRQPHRVVLDSPVR----TTGVQSPSP-----------PPSPPVPPPPPSPS 56
P +R SPP+ P R +P R T SP+P PP PP+P PPP P
Sbjct: 220 PRNRRSSPPKPPRRPTSSNPPRSARPTRSSPSPTPSPFLPNSPPLPPPPPPLPLPPPPPP 399
Query: 57 QVGSLERSPENSPAP 71
+ RS +S +P
Sbjct: 400 RPLRRRRSSRSSASP 444
Score = 36.6 bits (83), Expect = 0.074
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 6/62 (9%)
Frame = +1
Query: 20 PRQPHRVVLDSPVRTTG--VQSPSPP----PSPPVPPPPPSPSQVGSLERSPENSPAPEQ 73
P++P R SP+ G + SP PP SPP PP P+ S R +SP+P
Sbjct: 145 PKRPRRNPNPSPLPPPGSPISSPKPPRNRRSSPPKPPRRPTSSNPPRSARPTRSSPSPTP 324
Query: 74 QP 75
P
Sbjct: 325 SP 330
>AW620463
Length = 398
Score = 43.5 bits (101), Expect = 6e-04
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 2/60 (3%)
Frame = +3
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPE--NSPAPEQQP 75
+PP S +T P P P PP+PPPPP PS S SP ++P+P P
Sbjct: 159 NPPSSHGSATPASSKSSTTAPPPPPQPPPPLPPPPPPPSSPAS*PSSPSSPSTPSPSSPP 338
Score = 32.3 bits (72), Expect = 1.4
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 4/44 (9%)
Frame = +3
Query: 17 ISPPRQPHRVVLDSPVRTTGVQSPSP--PP--SPPVPPPPPSPS 56
+ PP P P + +PSP PP SPP PPP P+PS
Sbjct: 249 LPPPPPPPSSPAS*PSSPSSPSTPSPSSPPMTSPPTPPPGPAPS 380
>TC233861 weakly similar to UP|Q41192 (Q41192) NaPRP3, partial (21%)
Length = 438
Score = 42.7 bits (99), Expect = 0.001
Identities = 23/66 (34%), Positives = 30/66 (44%)
Frame = +2
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPPPSPPVPPPPPSPSQVGSLERSPENSPAPEQQPAV 77
SPP++P SP + + PS PPSPP PPPPP P S S ++
Sbjct: 65 SPPQRPWNWTSTSP-SASS*RKPSRPPSPPSPPPPPPPPPRSSNNPSSSSTMTTSTP*RC 241
Query: 78 TQEQWR 83
+ WR
Sbjct: 242 SARSWR 259
>TC207742 similar to UP|Q9FYG0 (Q9FYG0) F1N21.15 (At1g67330), partial (70%)
Length = 1115
Score = 42.4 bits (98), Expect = 0.001
Identities = 26/60 (43%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Frame = -2
Query: 18 SPPRQPHRVVLDSPVRTTGVQSPSPPP--SPPVPPPPPSPSQVGSLERSPENSPAPEQQP 75
SPP PHR + R SPSPPP SPP P PP + S RS N P P +P
Sbjct: 727 SPPSDPHRARTRASHRIPSCPSPSPPPRISPPCAPAPPQSTP--SARRS*SN-PTPSHKP 557
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,480,156
Number of Sequences: 63676
Number of extensions: 844051
Number of successful extensions: 33335
Number of sequences better than 10.0: 1148
Number of HSP's better than 10.0 without gapping: 10558
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18944
length of query: 1291
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1183
effective length of database: 5,762,624
effective search space: 6817184192
effective search space used: 6817184192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0331c.2


