

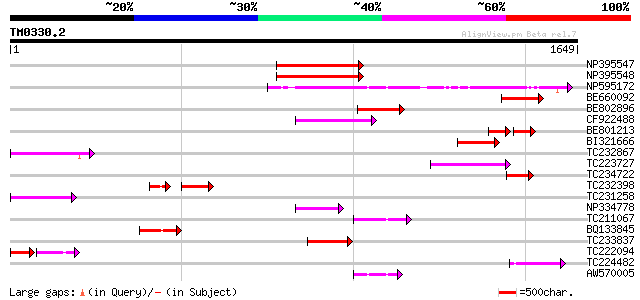
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0330.2
(1649 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP395547 reverse transcriptase [Glycine max] 417 e-116
NP395548 reverse transcriptase [Glycine max] 411 e-114
NP595172 polyprotein [Glycine max] 384 e-106
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 193 5e-49
BE802896 182 1e-45
CF922488 171 2e-42
BE801213 weakly similar to GP|6691193|gb| F7F22.17 {Arabidopsis ... 101 2e-41
BI321666 164 3e-40
TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial ... 139 1e-32
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 137 5e-32
TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (... 136 9e-32
TC232398 90 1e-28
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 121 2e-27
NP334778 reverse transcriptase [Glycine max] 116 9e-26
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 113 8e-25
BQ133845 112 1e-24
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 108 3e-23
TC222094 72 8e-21
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 92 2e-18
AW570005 80 6e-15
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 417 bits (1072), Expect = e-116
Identities = 188/254 (74%), Positives = 223/254 (87%)
Frame = +1
Query: 776 VRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGITVVANENNELIPTRQVTKWRVCIDY 835
VRKE+ KLL+AG+IYPISDS WVSPVQVVPKKGG+TVV N+ NELIPTR+VT+WR+CIDY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 836 RRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAFTCPYDV 895
R+LN TRKDH+PLPF+DQML +LA +Y FLDGYSGYNQI V P+DQEKTAFTCP+ V
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 896 FAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNLALVLKR 955
FAY+RMPFGLCNA TFQRCM AIF D++E CIE+FMDDFS FG +F CL NL VL+R
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 956 CQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGIRSFLGH 1015
C+++NLVLNWEKCHFMV++GIVLGHK+S+RGIEV + K++VI+KLPPP N+KGI SFLGH
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 1016 AGFYRRFIKDFSKL 1029
GFYRRFIKDF+K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 411 bits (1057), Expect = e-114
Identities = 187/254 (73%), Positives = 221/254 (86%)
Frame = +1
Query: 776 VRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGGITVVANENNELIPTRQVTKWRVCIDY 835
VRKE++KLL+ G+IYPISDS WVSPV VV KK G+TV+ NE N+LIPTR VT W++CIDY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 836 RRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAFTCPYDV 895
R+LN TRKDHFPLPF+DQML++LAGH YYCFLD Y GYNQI V P+DQEK AFTCP+ V
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 896 FAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNLALVLKR 955
FAY+R+PFGLCNAP TFQ CM AIF+D++E IE+FMDDFSVF P+ ++CL L +VL+R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 956 CQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGIRSFLGH 1015
C ETNLVLNWEKCHFMVR+GIVLGHK+S RGIEVD+ KI+VIEKLPPP+N+KGIRSFLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 1016 AGFYRRFIKDFSKL 1029
A FYRRFIKDF+K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 384 bits (986), Expect = e-106
Identities = 279/898 (31%), Positives = 434/898 (48%), Gaps = 12/898 (1%)
Frame = +1
Query: 750 HKILLEENYKPIVQPQRRLNPSMKDVVRKEIIKLLDAGVIYPISDSEWVSPVQVVPKKGG 809
H I L++ P+ R + KD + K I ++L G+I P S+S + P+ +V KK G
Sbjct: 1759 HAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQP-SNSPFSLPILLVKKKDG 1935
Query: 810 ITVVANENNELIPTRQVTKWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLD 869
WR C DYR LN++T KD FP+P +D++LD+L G QY+ LD
Sbjct: 1936 ------------------SWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLD 2061
Query: 870 GYSGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIE 929
SGY+QI V PED+EKTAF + + + MPFGL NAPATFQ M IF + +
Sbjct: 2062 LRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVL 2241
Query: 930 IFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEV 989
+F DD ++ ++ L +L VL+ ++ L KC F + LGHKVS G+ +
Sbjct: 2242 VFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSM 2421
Query: 990 DRAKIEVIEKLPPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENC 1049
+ K++ + P P N+K +R FLG G+YRRFIK ++ +A P+T+LL+K++ F ++
Sbjct: 2422 ENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKDS-FLWNNEA 2598
Query: 1050 LKAFESIKKSLVTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLN 1109
AF +KK++ APV+ PD+S PF + DAS + +GAVL Q + I Y S L
Sbjct: 2599 EAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQNG----HPIAYFSKKLA 2766
Query: 1110 EAQRNYTTTEKELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWVLL 1169
+ + +ELL + A KFR Y+LG K ++ TD +L+ L + P W+
Sbjct: 2767 PRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHK 2946
Query: 1170 LQEFDLEIIDRRGKDNGVADHLSRLEGGACSPIPIQEEFPDEKLLAVSTEEPLPWYVHFA 1229
+D +I + GKDN AD LSR + ++ EP ++
Sbjct: 2947 FLGYDFKIEYKPGKDNQAADALSR-------------------MFMLAWSEPHSIFLEEL 3069
Query: 1230 NFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVIRRCIPEVNFEKILWYCH 1289
R+ PH + K DA Y + L+ D V+ E+ KIL H
Sbjct: 3070 RARLIS-DPHLKQLMETYKQGADASHYTVREGLLY--WKDRVVIPAEEEI-VNKILQEYH 3237
Query: 1290 GSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNEMPLKNILE 1349
S GGH RT A+ L++ FYWP + D +A+++ C CQ+ N +P +
Sbjct: 3238 SSPIGGHAGITRTLAR-LKAQFYWPKMQEDVKAYIQKCLICQQA---KSNNTLPAGLLQP 3405
Query: 1350 IEL-FDVW---GIDFMGPFPPSFGCQYILLAVDYVSKWVEAAALSTN-DSKVVVAFLKKN 1404
+ + VW +DF+ P SFG I++ +D ++K+ L + +SKVV +
Sbjct: 3406 LPIPQQVWEDVAMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSH 3585
Query: 1405 IFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRIL 1464
I G+PR+I+SD F + ++ L + G +S+ YHPQ+ GQ E+ N+ L+ L
Sbjct: 3586 IVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYL 3765
Query: 1465 EKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKL 1524
K W + L A + Y TA+ +G +PF ++G+ P L +A
Sbjct: 3766 RCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGRE---PPTLTRQA------C 3918
Query: 1525 NFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREFVSGQLVLLFNS 1584
+ D + QL + D + + + ++ K+ D+K L+ F G VL+
Sbjct: 3919 SIDDPAEVRE---QLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQ 4089
Query: 1585 RLRLFPG------KLKSRWSGPFVVKRVFPHGAVEVENPETKNTFTV-NGQRLKVYHG 1635
R KL R+ GPF V A ++E P V + +LK ++G
Sbjct: 4090 PYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLELPSAARIHPVFHVSQLKPFNG 4263
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 193 bits (491), Expect = 5e-49
Identities = 88/123 (71%), Positives = 105/123 (84%)
Frame = -3
Query: 1430 SLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTA 1489
+LL+KYGV H+VSTPYHPQT+GQ EISNRE+KRILEK+V SRKDWS +LDDALWA+RTA
Sbjct: 370 ALLKKYGVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTA 191
Query: 1490 FKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRA 1549
+K PIG SP+ +VFGKACHLPVE+EHKAYWA++ NF A E+R LQL+ELDE RL A
Sbjct: 190 YKAPIGMSPYRVVFGKACHLPVEIEHKAYWAVKTCNFSMDQAGEERKLQLSELDEIRLEA 11
Query: 1550 YES 1552
YE+
Sbjct: 10 YEN 2
>BE802896
Length = 416
Score = 182 bits (462), Expect = 1e-45
Identities = 83/138 (60%), Positives = 110/138 (79%)
Frame = -2
Query: 1011 SFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVIVAPD 1070
SFLGHAGFYRRFI+DF K+A P++NLL+KE F F++ C AF+ +K++L+T P+I APD
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 1071 WSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEKELLGVVFACE 1130
W+ PFE+MCDAS+ ALG VL QK +++ VIYY+S L+ AQ NYTTTEKELL +VFA E
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 1131 KFRPYILGFKVVVHTDHA 1148
KF Y+LG +++V+ DHA
Sbjct: 55 KFHSYLLGTRIIVYIDHA 2
>CF922488
Length = 741
Score = 171 bits (434), Expect = 2e-42
Identities = 97/237 (40%), Positives = 135/237 (56%)
Frame = +3
Query: 831 VCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAFT 890
+C+DYR LN + KD FPLP I+ ++D + F+DG+SGYNQI +APED EKT F
Sbjct: 30 MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDMEKTTFI 209
Query: 891 CPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNLA 950
+ F YK M FGL N AT+QR M A+F D++ IE++MDD V + L NL
Sbjct: 210 TLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEEHLVNLR 389
Query: 951 LVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSERGIEVDRAKIEVIEKLPPPTNIKGIR 1010
+ +R ++ L LN KC F V+ +L S RGIEVD K++VI ++ P K ++
Sbjct: 390 KLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMAKPHTEKQVQ 569
Query: 1011 SFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSLVTAPVIV 1067
FLG + RFI +P+ LL K +D +C AFE IK+ L+ V+V
Sbjct: 570 GFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLINPHVLV 740
>BE801213 weakly similar to GP|6691193|gb| F7F22.17 {Arabidopsis thaliana},
partial (3%)
Length = 416
Score = 101 bits (252), Expect(2) = 2e-41
Identities = 46/63 (73%), Positives = 54/63 (85%)
Frame = +2
Query: 1465 EKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKL 1524
EK V SSRKDWS KL+DALWA +TA KTPIG +PF +V+ KACHLPVEL+HKAYWA++ L
Sbjct: 224 EKNVASSRKDWSSKLEDALWACKTAKKTPIGLTPFQMVYRKACHLPVELKHKAYWAMKCL 403
Query: 1525 NFD 1527
NFD
Sbjct: 404 NFD 412
Score = 88.2 bits (217), Expect(2) = 2e-41
Identities = 38/66 (57%), Positives = 51/66 (76%)
Frame = +3
Query: 1392 NDSKVVVAFLKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSG 1451
ND+K+V+ FLKKNIF+RFG+PR +ISDGG+HF +L+ V+HKV T YHPQT+G
Sbjct: 3 NDAKIVIKFLKKNIFSRFGMPRILISDGGSHFYYSQLNKVLKHDSVRHKVETSYHPQTNG 182
Query: 1452 QVEISN 1457
Q ++SN
Sbjct: 183 QAKVSN 200
>BI321666
Length = 430
Score = 164 bits (415), Expect = 3e-40
Identities = 71/123 (57%), Positives = 96/123 (77%)
Frame = +2
Query: 1303 AAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNEMPLKNILEIELFDVWGIDFMG 1362
+ VLQS FY P++ +D+ +SC++CQRT ++S+RNE+PL ILE+E+FD WGIDF+G
Sbjct: 5 STNVLQSRFYLPSIFKDAYVHAQSCNKCQRTRSVSKRNELPLHTILEVEIFDYWGIDFVG 184
Query: 1363 PFPPSFGCQYILLAVDYVSKWVEAAALSTNDSKVVVAFLKKNIFTRFGVPRAIISDGGTH 1422
PFPPSF +YIL+ VDYVSKWVEA A +D+K+V+ FLKK IF+R GVP +I +GG+H
Sbjct: 185 PFPPSFSNEYILVVVDYVSKWVEAVACQKSDAKIVIKFLKKQIFSRLGVPWVLIDNGGSH 364
Query: 1423 FCN 1425
CN
Sbjct: 365 LCN 373
>TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial (3%)
Length = 1144
Score = 139 bits (350), Expect = 1e-32
Identities = 85/255 (33%), Positives = 133/255 (51%), Gaps = 11/255 (4%)
Frame = -3
Query: 2 SEDPHAHMERFIRNCNTYRVMNVPSEAIRLSLFPFSLKDAAEDWLNSQPQGSLTTWEDLA 61
+EDP+AH+ +I CNT R+ VP++AIRLSL FSL A+ WL+S SL +W+++
Sbjct: 767 NEDPYAHLATYIEICNTIRLAGVPADAIRLSLLSFSLSGEAKRWLHSFKGNSLKSWDEVV 588
Query: 62 EKFTTRFFPRSLLRKLKNDIMTFAQSTDENLYEAWEHFKKLLRKCPQHNLTQAECVAKFY 121
EKF ++FP S + K I +F Q DE+L EA E F+ LLRK P H ++ + F
Sbjct: 587 EKFLKKYFPESKTAEGKAAISSFHQFPDESLSEALERFRGLLRKTPTHGFSEPIQLNIFI 408
Query: 122 DGLLYSSRFGLDAASSGEFDALSPQVGYNLIEKMAMRAMNSENER---QIRRSSFEVETY 178
D L S+ +DA+ G+ +P +LIE MA + +R ++S E+ +
Sbjct: 407 DELRPESKQLVDASVGGKIKMKTPDEAMDLIESMAASDIAILRDRAHIPTKKSLLELTSQ 228
Query: 179 DKLIASNKQLSEEVAEMRKHIKE--------TKSIGARVTCLECKFCGESHDSNQCTVND 230
D L+A NK LS+++ + K + + S + + C GE+H+S C N+
Sbjct: 227 DTLLAQNKLLSKQLETLTKTLSKLPTQLHSAQTSHSSILQVTGCTIFGEAHESGCCIPNE 48
Query: 231 DDSKVRTLTQGGKTP 245
+ G + P
Sbjct: 47 EQIAHEVNYMGNRPP 3
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 137 bits (344), Expect = 5e-32
Identities = 74/236 (31%), Positives = 119/236 (50%), Gaps = 2/236 (0%)
Frame = +1
Query: 1223 PWYVHFANFRVAGLIPHDLTWQQKKKFLHDAKSYLWDDPFLFKICSDGVIRRCIPEVNFE 1282
PWY + ++ ++ K+ A + L+K D RC+
Sbjct: 136 PWYFDIKRYVISKEYLPEIADNDKRTLRRLAAGFFMSGSILYKRNHDMKPLRCVDAREAN 315
Query: 1283 KILWYCHGSSYGGHFSGERTAAKVLQSGFYWPTLNRDSRAFVESCDRCQRTGNISRRNEM 1342
++ H S+G H +G A K+L++G+YW T+ D V C +CQ +
Sbjct: 316 HMIEEVHEGSFGTHANGHAMARKILRAGYYWLTMESDCCVHVRKCHKCQAFADNVNAPPH 495
Query: 1343 PLKNILEIELFDVWGIDFMGPFPP--SFGCQYILLAVDYVSKWVEAAALSTNDSKVVVAF 1400
PL + F +WGID +G P S G ++IL+A+DY +KWVEAA+ + VVV F
Sbjct: 496 PLNVMSSPWPFSMWGIDVIGAIEPKASNGHRFILVAIDYFTKWVEAASYTDVMRGVVVRF 675
Query: 1401 LKKNIFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEIS 1456
+KK I R+G+PR II+D GT+ N+ + E++ ++H TPY P+ + VE++
Sbjct: 676 IKKEIICRYGLPRKIITDNGTNLNNKMMGEICEEFKIQHHNPTPYRPKMN*AVEVA 843
>TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (32%)
Length = 482
Score = 136 bits (342), Expect = 9e-32
Identities = 59/78 (75%), Positives = 69/78 (87%)
Frame = -3
Query: 1445 YHPQTSGQVEISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFG 1504
YHPQT+GQ E+SN+E+KR+LE +V SSRKDW+ KLDDA WAYR AFKTPIG SPF LV+G
Sbjct: 234 YHPQTNGQAEVSNKEIKRVLENIVVSSRKDWALKLDDAFWAYRIAFKTPIGLSPFQLVYG 55
Query: 1505 KACHLPVELEHKAYWAIR 1522
KACHL VELEHKAYWA++
Sbjct: 54 KACHLSVELEHKAYWALK 1
>TC232398
Length = 1054
Score = 90.1 bits (222), Expect(2) = 1e-28
Identities = 38/93 (40%), Positives = 66/93 (70%)
Frame = +1
Query: 499 KRLNLGEVTPTMISLQMADRSLKTPYGIIEDVVVKVDKFVFPVDFVILDMEEDSKVPLIL 558
+R+ ++ PT ++LQ+AD S+ +G++ED++VKV + +F VDFVI+D+EED+++ LIL
Sbjct: 172 RRIGNQKIEPTRMTLQLADHSITRSFGVVEDILVKVHQLIFLVDFVIMDIEEDAEIRLIL 351
Query: 559 GRPFLATGEAEIKVAKGTLTLKVGEDEVLFNIF 591
G PF+ T + + + KG L + V + + FN+F
Sbjct: 352 GWPFMVTAKCVVDMGKGNLEMSVEDQKATFNLF 450
Score = 56.6 bits (135), Expect(2) = 1e-28
Identities = 30/60 (50%), Positives = 41/60 (68%)
Frame = +2
Query: 407 LHINIPFSEALEQMPIYAKFMKDILSKRRKLSEVDETILMTEECSAILQRKMPKKRRDPG 466
L I IPF E ++QMP+Y KF+KDIL K+ K ETI++ E C A++Q K+P K +D G
Sbjct: 2 LEITIPFGERIQQMPLYKKFLKDILIKKGKYIN-SETIVVGEYCRALIQ-KLPPKFKDLG 175
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 121 bits (304), Expect = 2e-27
Identities = 68/192 (35%), Positives = 98/192 (50%), Gaps = 2/192 (1%)
Frame = +2
Query: 3 EDPHAHMERFIRNCNTYRVMNVPSEAIRLSLFPFSLKDAAEDWLNSQPQGSLTTWEDLAE 62
EDPH H++ F C+T + +V + I L FP SL+ A+DWL S+ W+DL
Sbjct: 572 EDPHKHLKEFHIVCSTMKPPDVQEDHIFLKAFPHSLEGVAKDWLYYLAPRSIFNWDDLKR 751
Query: 63 KFTTRFFPRSLLRKLKNDIMTFAQSTDENLYEAWEHFKKLLRKCPQHNLTQAECVAKFYD 122
F +FFP S ++ DI Q + E+LYE WE FKK CP H +++ + FY+
Sbjct: 752 VFLEKFFPASRTTTIRKDISGIRQLSRESLYEYWERFKKSCASCPHHQISKQLLL*YFYE 931
Query: 123 GLLYSSRFGLDAASSGEFDALSPQVGYNLIEKMAMRA--MNSENERQIRRSSFEVETYDK 180
L R +DAAS G ++P NLI+KMA + ++ N+ + R EV T
Sbjct: 932 ELSNMKRSMIDAASGGALGDMTPTEARNLIKKMASNSQQFSARNDAIVLRGVHEVATDSS 1111
Query: 181 LIASNKQLSEEV 192
NK L E +
Sbjct: 1112SSTENKSLRENL 1147
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 116 bits (290), Expect = 9e-26
Identities = 60/141 (42%), Positives = 84/141 (59%)
Frame = +3
Query: 830 RVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAF 889
R+C+DYR LN + KD+FPLP ID ++ +A + F+DG+SGYNQI +APED EKT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 890 TCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSVFGPNFDACLGNL 949
+ F YK M FGL N AT+ R M A+F D++ IE ++D+ + L NL
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 950 ALVLKRCQETNLVLNWEKCHF 970
+ + ++ L LN KC F
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVF 425
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 113 bits (282), Expect = 8e-25
Identities = 62/167 (37%), Positives = 97/167 (57%)
Frame = +1
Query: 1001 PPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSL 1060
P ++ IRSF G A FYRRF+ +FS +A P+ L++K FT+ E +AF +K+ L
Sbjct: 97 PTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKL 276
Query: 1061 VTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEK 1120
APV+ PD+S FE+ CDAS + + AVL Q + I Y S L+ A NY T +K
Sbjct: 277 TKAPVLALPDFSKTFELECDASGVGVRAVLLQGG----HPIAYFSEKLHSATLNYPTYDK 444
Query: 1121 ELLGVVFACEKFRPYILGFKVVVHTDHAALRHLFAKQDSKPRLIRWV 1167
EL ++ A + + +++ + V+H+DH +L+++ K R +WV
Sbjct: 445 ELYALIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWV 585
>BQ133845
Length = 389
Score = 112 bits (280), Expect = 1e-24
Identities = 57/123 (46%), Positives = 85/123 (68%)
Frame = +2
Query: 378 ELRKIPFPKALVKKNLDK*FSKFLEVFKKLHINIPFSEALEQMPIYAKFMKDILSKRRKL 437
E +++P+P K+ ++ +KFL++FKKL I +PF EAL+QMP+YA F+KD+L+K+
Sbjct: 23 ECKEVPYPLVPS*KDKEQHLAKFLDIFKKLEITLPFEEALQQMPLYANFLKDMLTKKNWY 202
Query: 438 SEVDETILMTEECSAILQRKMPKKRRDPGSFTIPVEIEGLTVVEALCDLGASINLMPLSM 497
D+ I++ CSA++QR +P DPG T+P I + V +AL DLGASINLMPLSM
Sbjct: 203 IHSDK-IVVEGNCSAVIQRILPP*HTDPGFVTMPCSIGEVAVGKALIDLGASINLMPLSM 379
Query: 498 FKR 500
++
Sbjct: 380 CRQ 388
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 108 bits (269), Expect = 3e-23
Identities = 57/133 (42%), Positives = 81/133 (60%)
Frame = +2
Query: 865 YCFLDGYSGYNQICVAPEDQEKTAFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLI 924
+ F+DG+SGYNQI +A ED EKT F + F+Y+ M FGL N AT+QR M A+F D++
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 925 ETCIEIFMDDFSVFGPNFDACLGNLALVLKRCQETNLVLNWEKCHFMVRDGIVLGHKVSE 984
IE+++DD L NL + R Q+ L LN KC F V+ G +LG VS+
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 985 RGIEVDRAKIEVI 997
+GIE+D K++ +
Sbjct: 362 KGIEIDPEKVKAL 400
>TC222094
Length = 984
Score = 72.4 bits (176), Expect(2) = 8e-21
Identities = 32/71 (45%), Positives = 44/71 (61%)
Frame = -2
Query: 2 SEDPHAHMERFIRNCNTYRVMNVPSEAIRLSLFPFSLKDAAEDWLNSQPQGSLTTWEDLA 61
+EDP+AH+ +I CNT +++ VP +AI L LF FSL A WL S +L TW +
Sbjct: 707 TEDPYAHLATYIDICNTVKIVGVPEDAIHLDLFCFSLAGEARTWLRSFKGNNLRTWNEXX 528
Query: 62 EKFTTRFFPRS 72
EKF ++FP S
Sbjct: 527 EKFLKKYFPES 495
Score = 48.1 bits (113), Expect(2) = 8e-21
Identities = 38/125 (30%), Positives = 58/125 (46%)
Frame = -3
Query: 78 KNDIMTFAQSTDENLYEAWEHFKKLLRKCPQHNLTQAECVAKFYDGLLYSSRFGLDAASS 137
K +I +F Q E+L EA + F LL K P H ++ + F DG+ S+ LDA++
Sbjct: 478 KVEISSFHQHPHESLSEALDRFHGLLWKTPTHGFSEPVQLNIFIDGMQPHSKQLLDASAG 299
Query: 138 GEFDALSPQVGYNLIEKMAMRAMNSENERQIRRSSFEVETYDKLIASNKQLSEEVAEMRK 197
G+ +P+ LIE MA N+ I R E ++ + L + + E R
Sbjct: 298 GKIKLKTPEEAIELIENMA------ANDYVILRDQ-EPSPQEESTRLEELLVQFMQETRS 140
Query: 198 HIKET 202
H K T
Sbjct: 139 HQKST 125
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 92.0 bits (227), Expect = 2e-18
Identities = 56/164 (34%), Positives = 88/164 (53%), Gaps = 1/164 (0%)
Frame = +1
Query: 1454 EISNRELKRILEKVVDSSRKDWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVEL 1513
E +N+ +K+I++K+ S KDW L AL YRT+ +T G +PF LV+G LP E+
Sbjct: 1 EAANKNIKKIIQKMT-VSYKDWHEMLPFALHGYRTSVRTSTGATPFSLVYGMEAVLPFEV 177
Query: 1514 EHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNREF 1573
E + + + ++ R QLN ++ RL A +Y+++ K D+K+ R+F
Sbjct: 178 EVPSLRILAESGLKESEWAQTRYDQLNLIEGKRLTAMSHGRLYQQRMKSAFDKKVCLRKF 357
Query: 1574 VSGQLVL-LFNSRLRLFPGKLKSRWSGPFVVKRVFPHGAVEVEN 1616
G LVL + ++ GK + GPFVVKR F GA+ + N
Sbjct: 358 HEGDLVLKKMSHAVKDHRGKWAPNYEGPFVVKRAFSGGALVLTN 489
>AW570005
Length = 413
Score = 80.5 bits (197), Expect = 6e-15
Identities = 46/142 (32%), Positives = 79/142 (55%)
Frame = -2
Query: 1001 PPPTNIKGIRSFLGHAGFYRRFIKDFSKLAKPMTNLLEKEAPFTFDENCLKAFESIKKSL 1060
PPP + +R FL GFYRRFIK ++ +A P+++LL K++ F + AF+++K +
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDS-FVWSPEADVAFQALKNVV 236
Query: 1061 VTAPVIVAPDWSLPFEIMCDASDLALGAVLCQKKERVLYVIYYASTVLNEAQRNYTTTEK 1120
V+ PD++ PF + DAS +GAVL Q+ + +++ + R+ +T
Sbjct: 235 TNTLVLALPDFTKPFTVETDASGSDMGAVLSQEGHP---IAFFSKEFCPKLVRS-STYVH 68
Query: 1121 ELLGVVFACEKFRPYILGFKVV 1142
EL + +K+R Y+LG +V
Sbjct: 67 ELAAITNVVKKWRQYLLGHHLV 2
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 73,583,288
Number of Sequences: 63676
Number of extensions: 1064199
Number of successful extensions: 4679
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 4615
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4663
length of query: 1649
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1539
effective length of database: 5,635,272
effective search space: 8672683608
effective search space used: 8672683608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0330.2


