

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0327.6
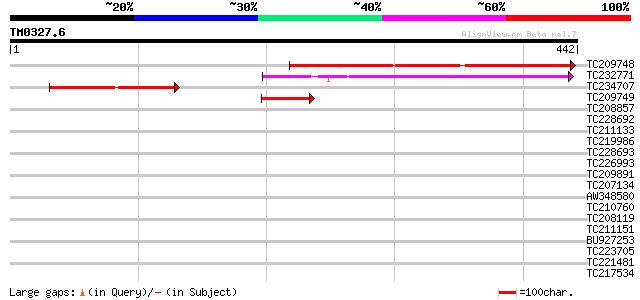
(442 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209748 similar to PIR|D88042|D88042 protein F56D12.5 [imported... 355 2e-98
TC232771 147 9e-36
TC234707 weakly similar to UP|P91672 (P91672) FGF homolog, parti... 122 2e-28
TC209749 82 6e-16
TC208857 weakly similar to UP|Q84KG8 (Q84KG8) Fatty acid delta-6... 36 0.030
TC228692 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, pa... 35 0.088
TC211133 weakly similar to UP|Q9BKV7 (Q9BKV7) Ppg3, partial (4%) 34 0.12
TC219986 weakly similar to UP|Q41187 (Q41187) Glycine-rich prote... 33 0.34
TC228693 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, pa... 32 0.44
TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 deh... 32 0.75
TC209891 weakly similar to UP|Q9LW52 (Q9LW52) Arabidopsis thalia... 32 0.75
TC207134 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 31 0.97
AW348580 31 1.3
TC210760 weakly similar to UP|Q9EPR8 (Q9EPR8) Cdk3-binding prote... 31 1.3
TC208119 similar to GB|AAL25838.1|23379334|AY057393 one-helix pr... 30 2.2
TC211151 UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, complete 30 2.2
BU927253 similar to GP|14194119|gb| At1g01540/F22L4_6 {Arabidops... 30 2.8
TC223705 30 2.8
TC221481 30 2.8
TC217534 similar to GB|AAB62826.1|2252827|IG005I10 coded for by ... 30 2.8
>TC209748 similar to PIR|D88042|D88042 protein F56D12.5 [imported] -
Caenorhabditis elegans {Caenorhabditis elegans;} ,
partial (5%)
Length = 894
Score = 355 bits (912), Expect = 2e-98
Identities = 174/223 (78%), Positives = 194/223 (86%)
Frame = +1
Query: 219 QVRARVDNIRLHVARLGFNKDGDDVDFIDEKHFPSRVRVWVGPEIGAAYVGGLSLGRSTE 278
QVRARVDNIRLH+ARL +++ D DF++EKH SRVRVWVGPEIG+ Y GLSLGRSTE
Sbjct: 1 QVRARVDNIRLHLARLXXSQNDDVDDFVEEKHXXSRVRVWVGPEIGSTYCAGLSLGRSTE 180
Query: 279 NNEREVEIQKIVKGNFEKSEHISKVKAAARSSRRTRTKSWRMDQDAEGNAAIFDVVLHDN 338
N E EVE ++IV+GNFEKS+ S VKA A+SSRRTR++SWRMDQDAEGNAAIFDVVLHDN
Sbjct: 181 NKECEVETKRIVEGNFEKSQG-SNVKALAKSSRRTRSRSWRMDQDAEGNAAIFDVVLHDN 357
Query: 339 MTGLEVGAWKPTGQTGDDPVHGMRGRYSGASRPFNKSGSVVIAGDEYGEEVGWRLSKEME 398
TG EVG+W+P G DDP HG+RGRY A+R FNKSGSVVIAGDEYGEEVGWRLSKEME
Sbjct: 358 TTGQEVGSWRPKG---DDPAHGLRGRYVRANRAFNKSGSVVIAGDEYGEEVGWRLSKEME 528
Query: 399 GSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEWCDEVDLPLIN 441
GSVLKWRIGGEFWVSY PNQ KGSYFETRY+EWCDEVDLPLI+
Sbjct: 529 GSVLKWRIGGEFWVSYLPNQAKGSYFETRYIEWCDEVDLPLIH 657
>TC232771
Length = 1027
Score = 147 bits (371), Expect = 9e-36
Identities = 84/248 (33%), Positives = 131/248 (51%), Gaps = 6/248 (2%)
Frame = +3
Query: 198 HHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFNKDGDDVDFID------EKHF 251
+ QLE +Q Y V + ++++ +DNIR V +L +D+ + EKHF
Sbjct: 93 YQQLEGVLQFNYKVTIRERWVEIVVNIDNIRCDVFKLV-----NDILMRERGAGAAEKHF 257
Query: 252 PSRVRVWVGPEIGAAYVGGLSLGRSTENNEREVEIQKIVKGNFEKSEHISKVKAAARSSR 311
PSR+ + + P I V +S+G+S+EN E ++K ++ +FE + +A S
Sbjct: 258 PSRISLQLTPTI-QQQVLTVSVGKSSENPRIEFGVEKGIEASFEPPNPYIGLSVSAGEST 434
Query: 312 RTRTKSWRMDQDAEGNAAIFDVVLHDNMTGLEVGAWKPTGQTGDDPVHGMRGRYSGASRP 371
K W+ +Q G +A + LHD+ G EV + KP+ +P + RYS A RP
Sbjct: 435 TVSLKPWKFEQSVHGYSANLNWFLHDSTDGKEVFSTKPSKVALFNPKSWFKDRYSSAYRP 614
Query: 372 FNKSGSVVIAGDEYGEEVGWRLSKEMEGSVLKWRIGGEFWVSYWPNQVKGSYFETRYVEW 431
F + G V+ AGDEYGE V W++ K G ++W I G W++YWPN+ K Y ETR E+
Sbjct: 615 FTRQGGVIFAGDEYGESVCWKVDKGAVGKTMEWEIRGWIWITYWPNKHKTFYHETRRFEF 794
Query: 432 CDEVDLPL 439
+ V L +
Sbjct: 795 RETVYLKI 818
>TC234707 weakly similar to UP|P91672 (P91672) FGF homolog, partial (3%)
Length = 324
Score = 122 bits (307), Expect = 2e-28
Identities = 61/101 (60%), Positives = 73/101 (71%)
Frame = +1
Query: 32 QLNSLNRTQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSA 91
QLNS + +S ANLR+RL GD+HR+LARL P Q +AS+G+R++ P R K +P S
Sbjct: 28 QLNSFHSPES*ANLRLRLGSCGDLHRMLARLSPTQRSQASMGLRKVPPLIR--KAVPPSP 201
Query: 92 PPTPPGNHIPLTHRAR*HLPKIPAPETEAGTHIMDHGLTHT 132
PPTPPGNHIPL H AR*HLP PAPET+ H MDHGL HT
Sbjct: 202 PPTPPGNHIPLPHGAR*HLPPFPAPETKTRPHRMDHGLAHT 324
>TC209749
Length = 460
Score = 81.6 bits (200), Expect = 6e-16
Identities = 37/41 (90%), Positives = 41/41 (99%)
Frame = +3
Query: 197 AHHQLEAHVQLEYTVGFHDGFIQVRARVDNIRLHVARLGFN 237
AHHQLEAHVQL+YTVGF+DGFIQVRARVDNIRLH+ARLGF+
Sbjct: 336 AHHQLEAHVQLDYTVGFYDGFIQVRARVDNIRLHLARLGFS 458
Score = 35.0 bits (79), Expect = 0.068
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 4/36 (11%)
Frame = +2
Query: 170 SHSRDGVVED----PQNVLPHRRSRHRALFHAHHQL 201
+ S +GVV+ P N L HRR RHR L HAH +L
Sbjct: 2 ARSPNGVVQTRILRPPNFLHHRRRRHRTLLHAHPRL 109
>TC208857 weakly similar to UP|Q84KG8 (Q84KG8) Fatty acid delta-6 desaturase,
partial (21%)
Length = 648
Score = 36.2 bits (82), Expect = 0.030
Identities = 27/83 (32%), Positives = 34/83 (40%), Gaps = 6/83 (7%)
Frame = +2
Query: 46 RVRLRRGGDIHRVLA------RLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNH 99
R+ L D+ R+L R PP HP+ S P P PSS+PP+PP
Sbjct: 104 RLDLHPRQDLRRLLLAPPPPRRPPPTPHPRRERTPPTPSSPSTLPTPPPSSSPPSPPPTV 283
Query: 100 IPLTHRAR*HLPKIPAPETEAGT 122
P T P P P T A +
Sbjct: 284 FPTT-------PSPPPPPTTASS 331
>TC228692 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, partial (48%)
Length = 775
Score = 34.7 bits (78), Expect = 0.088
Identities = 26/89 (29%), Positives = 34/89 (37%), Gaps = 3/89 (3%)
Frame = +2
Query: 46 RVRLRRGGDIHRVLARLPPIQH-PKASLGIREMQPFSR*P--KPLPSSAPPTPPGNHIPL 102
R+ LRR IHR R P+ H P + +P S P + LP PP PP +
Sbjct: 236 RLHLRRHRQIHRRCHRRHPLHHLPPHPSDLHPHRPPSHGPHLRALPRHRPPPPPHPQL-- 409
Query: 103 THRAR*HLPKIPAPETEAGTHIMDHGLTH 131
HLP + G H + H
Sbjct: 410 ------HLPNLKPQSNSLGLHELQRRTCH 478
>TC211133 weakly similar to UP|Q9BKV7 (Q9BKV7) Ppg3, partial (4%)
Length = 709
Score = 34.3 bits (77), Expect = 0.12
Identities = 31/93 (33%), Positives = 41/93 (43%), Gaps = 2/93 (2%)
Frame = +2
Query: 58 VLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIPLTHR--AR*HLPKIPA 115
+L RLPP+ P L + P P P P PP PP +PL R R +P +PA
Sbjct: 161 LLRRLPPLLLPLLHLQL----PLPPPPLPPPLLPPPLPP-LLLPLPRRLLLRLPIPPLPA 325
Query: 116 PETEAGTHIMDHGLTHTGISLQFLQPRLHRAPL 148
P + HG+ +L+ L R APL
Sbjct: 326 PGMQGPQE--GHGVRFADSALRLLLRRNR*APL 418
>TC219986 weakly similar to UP|Q41187 (Q41187) Glycine-rich protein
(Fragment), partial (47%)
Length = 584
Score = 32.7 bits (73), Expect = 0.34
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 5/58 (8%)
Frame = -3
Query: 84 PKPLPSSAPPTPPGNHIPLTHRAR*HLPKIPAP-----ETEAGTHIMDHGLTHTGISL 136
P+PLP +PPTPP P + P P P T + THI D T G S+
Sbjct: 174 PRPLPPPSPPTPPLPAPPPYPPPKPLSPPAPPP*PTS*TTTSSTHISDSSNTTDGPSI 1
>TC228693 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, partial (59%)
Length = 907
Score = 32.3 bits (72), Expect = 0.44
Identities = 28/89 (31%), Positives = 36/89 (39%), Gaps = 3/89 (3%)
Frame = +2
Query: 46 RVRLRRGGDIHRVLARLPPIQH-PKASLGIREMQPFSR*P--KPLPSSAPPTPPGNHIPL 102
R+ LRR IHR RL P+ H P +R + S P + LP PP P +H L
Sbjct: 227 RLHLRRHRQIHRRRHRLHPLHHLPPHPPDLRPHRSPSHGPHLRALPRHRPPPP--SHPQL 400
Query: 103 THRAR*HLPKIPAPETEAGTHIMDHGLTH 131
HLP + G H + H
Sbjct: 401 ------HLPNLKPQSNSLGLHELQRRTCH 469
>TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (94%)
Length = 1388
Score = 31.6 bits (70), Expect = 0.75
Identities = 30/89 (33%), Positives = 34/89 (37%), Gaps = 2/89 (2%)
Frame = +2
Query: 79 PFSR*PKPLPSS-APPTPPGNHIPLTHRAR*HLPKIPAPETEAGTHIMDHGLTHTGISLQ 137
P R PKP PS+ APPTPP N P P A T + T + +
Sbjct: 563 PLRRSPKPPPSTPAPPTPPPNAPPT------GTP*TTARLTPSSPATASSSTTSPPAAAR 724
Query: 138 FLQPRLHRAPLLALRL-*RAQRSWVTLLP 165
P R P A R RA SW T P
Sbjct: 725 IS*PARSRGPWAASRSGSRASSSWATSRP 811
>TC209891 weakly similar to UP|Q9LW52 (Q9LW52) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MLM24, partial (10%)
Length = 646
Score = 31.6 bits (70), Expect = 0.75
Identities = 22/70 (31%), Positives = 34/70 (48%), Gaps = 8/70 (11%)
Frame = -3
Query: 64 PIQHP-KASLGIREMQPFSR*PKPLP---SSAPPTPPGNHIPLTHRAR*HLP----KIPA 115
P QHP K +LG++ +P P P+P + PP PP P + A +P K+P
Sbjct: 368 PKQHPLKQNLGLKNPRPGRNPPPPMPPPGKNPPPPPPPMPPPGKYPAPPPMPPPGKKLPP 189
Query: 116 PETEAGTHIM 125
P + A ++
Sbjct: 188 PNSLASPPLL 159
>TC207134 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(67%)
Length = 1418
Score = 31.2 bits (69), Expect = 0.97
Identities = 30/95 (31%), Positives = 36/95 (37%), Gaps = 26/95 (27%)
Frame = +3
Query: 85 KPLPSSAPPTPPGNHIPLTHRAR*H--------------LPKIPAPETEAG--------- 121
+PLP + PPTPPG HR+R LP+ P P + +G
Sbjct: 9 RPLPKTRPPTPPGETPNPHHRSRPKRRAVARQALRRRFLLPR-PPPRSTSGRPLRHAEPP 185
Query: 122 --THIMDHGLTHTGISLQFLQ-PRLHRAPLLALRL 153
I L H S Q LQ P HR L RL
Sbjct: 186 VLRGIQHRRLRHPSRSFQNLQPPARHRLGLFQFRL 290
>AW348580
Length = 726
Score = 30.8 bits (68), Expect = 1.3
Identities = 26/88 (29%), Positives = 34/88 (38%)
Frame = +1
Query: 63 PPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIPLTHRAR*HLPKIPAPETEAGT 122
PP HP + + P S P PS +P +PP P P+ P+P T
Sbjct: 241 PPNPHPPPPPPLSSLSPISSSPSAWPSHSPSSPPFATPP--------PPRPPSPPP*TST 396
Query: 123 HIMDHGLTHTGISLQFLQPRLHRAPLLA 150
HT SL P L R+P+ A
Sbjct: 397 R---RDSVHT--SLPHNSPELTRSPVRA 465
>TC210760 weakly similar to UP|Q9EPR8 (Q9EPR8) Cdk3-binding protein ik3-1,
partial (4%)
Length = 826
Score = 30.8 bits (68), Expect = 1.3
Identities = 25/68 (36%), Positives = 30/68 (43%), Gaps = 7/68 (10%)
Frame = +1
Query: 64 PIQHPKASLGIREMQPFSR*PKPLPS-SAPPTPP------GNHIPLTHRAR*HLPKIPAP 116
P+Q P SL Q F P P PS S P PP G+H P+ R LP+ P
Sbjct: 232 PLQDPSRSL----QQHF---PLPHPSHSHRPNPPLRSRFNGSHHPMPIRRPRSLPRAPRA 390
Query: 117 ETEAGTHI 124
E G H+
Sbjct: 391 RDEPGGHV 414
>TC208119 similar to GB|AAL25838.1|23379334|AY057393 one-helix protein
{Arabidopsis thaliana;} , partial (49%)
Length = 662
Score = 30.0 bits (66), Expect = 2.2
Identities = 23/66 (34%), Positives = 33/66 (49%), Gaps = 4/66 (6%)
Frame = +3
Query: 35 SLNRTQSRANLRVRLRRG---GDIHRVLARLPPIQHPKASLGIREMQ-PFSR*PKPLPSS 90
SL + S+ + V +R G I R +A PPI+ P +S ++Q P P PSS
Sbjct: 42 SLRFSSSKPHNSVTIRNSQAEGPIRRPVA--PPIREPSSSSSAPQLQKPTLPSQPPSPSS 215
Query: 91 APPTPP 96
+PP P
Sbjct: 216 SPPQKP 233
>TC211151 UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, complete
Length = 1107
Score = 30.0 bits (66), Expect = 2.2
Identities = 15/41 (36%), Positives = 22/41 (53%)
Frame = +3
Query: 61 RLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIP 101
R I H KAS+G + + +R P +PPTPP + +P
Sbjct: 138 RFGAIPHQKASIGTKCSRSRARHRTPRRRFSPPTPPSSLLP 260
>BU927253 similar to GP|14194119|gb| At1g01540/F22L4_6 {Arabidopsis
thaliana}, partial (13%)
Length = 309
Score = 29.6 bits (65), Expect = 2.8
Identities = 17/46 (36%), Positives = 21/46 (44%)
Frame = +1
Query: 56 HRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGNHIP 101
HR R+P HP L + + P P P P PP PP +H P
Sbjct: 133 HRHPRRIPHRHHPLPPLPLPHLPP-PPPPPPPP*EPPPPPPLHHPP 267
>TC223705
Length = 918
Score = 29.6 bits (65), Expect = 2.8
Identities = 15/51 (29%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Frame = -2
Query: 54 DIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGN--HIPL 102
+IH+ +P + H G ++ P +R*P P+ PP P H+P+
Sbjct: 503 NIHQPTPFMPTLHHHLPHTGHMQIVPLAR*PIAPPTPLPPAPMARRAHLPV 351
>TC221481
Length = 516
Score = 29.6 bits (65), Expect = 2.8
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Frame = -2
Query: 177 VEDPQNVLPHRRSRHRALFHAHHQLEA-HVQLEYTVGFH 214
+E+ +LPH H L HHQ +A H+ L Y GFH
Sbjct: 299 MENDLYLLPHHNYHHHRLQGFHHQNKAVHLFLLYLHGFH 183
>TC217534 similar to GB|AAB62826.1|2252827|IG005I10 coded for by A. thaliana
cDNA N96903 {Arabidopsis thaliana;} , partial (98%)
Length = 1147
Score = 29.6 bits (65), Expect = 2.8
Identities = 23/73 (31%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Frame = +1
Query: 39 TQSRANLRVRLRRGGDIHRVLARLPPIQHPKASLGIREMQPFSR*PKPLPSSAPPTPPGN 98
T+ +L V R +H PP L +R+ + R P+P PSSA P P
Sbjct: 124 TERSHHLHVAPLRLHPLHHKGLLQPPSPRTHCPLHLRDARHQPRHPRPPPSSAFPRHPRQ 303
Query: 99 HIPL--THRAR*H 109
+ L HR+R H
Sbjct: 304 PLRLQPPHRSRPH 342
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,924,454
Number of Sequences: 63676
Number of extensions: 362219
Number of successful extensions: 4470
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 3664
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4204
length of query: 442
length of database: 12,639,632
effective HSP length: 100
effective length of query: 342
effective length of database: 6,272,032
effective search space: 2145034944
effective search space used: 2145034944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0327.6


