

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
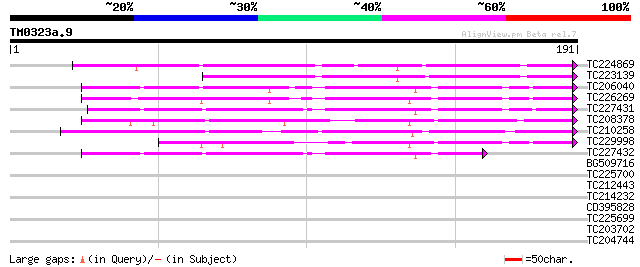
Query= TM0323a.9
(191 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224869 weakly similar to UP|ST14_SOLTU (Q41495) STS14 protein ... 112 1e-25
TC223139 weakly similar to UP|Q8LDA3 (Q8LDA3) Sts14, partial (59%) 100 3e-22
TC206040 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (... 77 5e-15
TC226269 homologue to UP|Q9XFB4 (Q9XFB4) PR1a precursor, complete 71 3e-13
TC227431 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (... 68 2e-12
TC208378 similar to UP|PR1_MEDTR (Q40374) Pathogenesis-related p... 68 2e-12
TC210258 weakly similar to UP|Q9LPM7 (Q9LPM7) F2J10.6 protein, p... 67 4e-12
TC229998 weakly similar to PIR|T04232|T04232 pathogenesis-relate... 67 7e-12
TC227432 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (... 51 4e-07
BG509716 similar to GP|4928711|gb|A PR1a precursor {Glycine max}... 31 0.40
TC225700 similar to UP|V722_ARATH (P47192) Vesicle-associated me... 28 3.4
TC212443 27 4.4
TC214232 weakly similar to UP|ATF5_HUMAN (Q9Y2D1) Cyclic-AMP-dep... 27 5.8
CD395828 similar to PIR|T51584|T515 splicing factor 9G8-like SR ... 27 5.8
TC225699 similar to UP|V722_ARATH (P47192) Vesicle-associated me... 26 9.8
TC203702 weakly similar to GB|AAP68215.1|31711718|BT008776 At1g5... 26 9.8
TC204744 homologue to UP|Q9MB06 (Q9MB06) Type 2A protein phospha... 26 9.8
>TC224869 weakly similar to UP|ST14_SOLTU (Q41495) STS14 protein precursor,
partial (65%)
Length = 1276
Score = 112 bits (280), Expect = 1e-25
Identities = 69/177 (38%), Positives = 96/177 (53%), Gaps = 7/177 (3%)
Frame = +3
Query: 22 HPFFAVLVALALLLVSSQAT------PATTDGAKVYLFEHNFVRREKGISEHLEWSDKLA 75
+P F++ L+++ AT P T A+ +L HN R E G+ E L WS+KL
Sbjct: 576 YPLFSLASLATFLVLTHAATAPENPPPPLTAAAREFLEAHNQARAEVGV-EALSWSEKLG 752
Query: 76 KDASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDR-FYYEG 134
+SL R++R+K C RYG NQ ++ PR V+ +WVK++ FY
Sbjct: 753 NVSSLMVRYQRNKKGCEFANLTASRYGG--NQLWAGVTEVA-PRVVVEEWVKEKKFYVRE 923
Query: 135 NNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
NN+CV + CG YTQ+VWR + +GCAQA C + L +CFY PPGNV GE P
Sbjct: 924 NNTCV-GKHECGVYTQVVWRNSTEVGCAQAVCVKEQAS--LTICFYDPPGNVIGEIP 1085
>TC223139 weakly similar to UP|Q8LDA3 (Q8LDA3) Sts14, partial (59%)
Length = 634
Score = 100 bits (250), Expect = 3e-22
Identities = 55/127 (43%), Positives = 72/127 (56%), Gaps = 1/127 (0%)
Frame = +2
Query: 66 EHLEWSDKLAKDASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQW 125
E L WS+++A S AR++R K C +YG NQ L + PR + +W
Sbjct: 2 EPLRWSEQVANVTSKLARYQRVKTGCQFANLTAGKYGA--NQLLARGSAAVTPRMAVEEW 175
Query: 126 VKDR-FYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPG 184
VK + FY+ +NSC P N+ CG YTQ+VWRK+ LGCAQA C + L +CFY PPG
Sbjct: 176 VKQKQFYFHADNSCAP-NHRCGVYTQVVWRKSVELGCAQATCVKEQAS--LTICFYNPPG 346
Query: 185 NVPGERP 191
N GE P
Sbjct: 347 NYVGESP 367
>TC206040 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (90%)
Length = 772
Score = 77.0 bits (188), Expect = 5e-15
Identities = 60/169 (35%), Positives = 84/169 (49%), Gaps = 2/169 (1%)
Frame = +3
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F VL L L+++ S A D Y+ HN R E G+ ++L W D +A A YA
Sbjct: 108 FPVLCVLGLVMIVSHVANAQ-DSPADYVNAHNAARSEVGV-QNLAWDDTVAAFAQNYANQ 281
Query: 85 RR-DKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPAN 142
R+ D L G +YG + L + + + ++ WV ++ Y+ N NSCV
Sbjct: 282 RKGDCQLIHSG--GGGQYG----ENLAMSTGDLSGTDAVKLWVDEKSNYDYNSNSCVGGE 443
Query: 143 YPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
C YTQ+VWR + LGCA+ C D GG + C Y PPGN G+RP
Sbjct: 444 --CLHYTQVVWRDSVRLGCAKVAC--DNGGTFI-TCNYAPPGNYVGQRP 575
>TC226269 homologue to UP|Q9XFB4 (Q9XFB4) PR1a precursor, complete
Length = 771
Score = 70.9 bits (172), Expect = 3e-13
Identities = 59/177 (33%), Positives = 81/177 (45%), Gaps = 10/177 (5%)
Frame = +3
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKG--------ISEHLEWSDKLAK 76
F V+ L L++V A D A+ Y+ HN R E G I L W D +A
Sbjct: 108 FCVMCVLGLVIVGDVAY--AQDSAEDYVNAHNAARAEVGSQSPRQTVIVPSLAWDDTVAA 281
Query: 77 DASLYARFRR-DKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-G 134
A YA R+ D L G YG + + + + + ++ WV ++ Y+
Sbjct: 282 YAESYANQRKGDCQLIHSG----GEYG----ENIAMSTGELSGTDAVKMWVDEKSNYDYD 437
Query: 135 NNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
+NSCV C YTQ+VW + LGCA+ C D GG + C Y PPGN GERP
Sbjct: 438 SNSCVGGE--CLHYTQVVWANSVRLGCAKVTC--DNGGTFI-TCNYDPPGNFVGERP 593
>TC227431 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (90%)
Length = 707
Score = 68.2 bits (165), Expect = 2e-12
Identities = 53/166 (31%), Positives = 79/166 (46%), Gaps = 1/166 (0%)
Frame = +2
Query: 27 VLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRR 86
+L L L++V A D Y+ HN R + G+ ++ W + +A A YA R+
Sbjct: 86 LLCVLGLVIVGDHVAYAQ-DSPTDYVNAHNAARSQVGVP-NIVWDNAVAAFAQNYANQRK 259
Query: 87 DKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPANYPC 145
D K YG + L + + ++ ++ WV ++ Y N NSCV C
Sbjct: 260 GDCKLVHSGGDGK-YG----ENLAGSTGNLSGKDAVQLWVNEKSKYNYNSNSCVGGE--C 418
Query: 146 GDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
YTQ+VWR + LGCA+ C + GG + C Y PPGN G+RP
Sbjct: 419 LHYTQVVWRNSLRLGCAKVRC--NNGGTFIG-CNYAPPGNYIGQRP 547
>TC208378 similar to UP|PR1_MEDTR (Q40374) Pathogenesis-related protein PR-1
precursor, partial (84%)
Length = 779
Score = 68.2 bits (165), Expect = 2e-12
Identities = 58/178 (32%), Positives = 75/178 (41%), Gaps = 11/178 (6%)
Frame = +1
Query: 25 FAVLVALALLLVSSQ---ATPATTDG-----AKVYLFEHNFVRREKGISEHLEWSDKLAK 76
F AL LLLV++ P TT A +L N R + L W KLA
Sbjct: 61 FLSCFALFLLLVATTYATVVPTTTQKPPRSFANQFLIPQNAARAVLRLRP-LVWDSKLAH 237
Query: 77 DASLYARFRRDKYLC--TPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE- 133
A YA RR+ + G + + + W P + + WV++R +Y
Sbjct: 238 YAQWYANQRRNDCALEHSNGPYGENIFWGSGTGW--------KPAQAVSAWVEERQWYNY 393
Query: 134 GNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
+NSC CG YTQIVW T+ +GCA C G C Y PPGN GERP
Sbjct: 394 WHNSCANGQM-CGHYTQIVWSTTRKIGCASVVCSGGKG--TFMTCNYDPPGNYYGERP 558
>TC210258 weakly similar to UP|Q9LPM7 (Q9LPM7) F2J10.6 protein, partial (66%)
Length = 608
Score = 67.4 bits (163), Expect = 4e-12
Identities = 52/175 (29%), Positives = 80/175 (45%), Gaps = 1/175 (0%)
Frame = +1
Query: 18 KMFPHPFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKD 77
K P +L L+ ++ + + +L HN R E G+ L W+ L
Sbjct: 46 KKMMSPSHVILSIFFLVCTTTPPLSLAQNTPQDFLDVHNQARAEVGVGP-LSWNHTLQAY 222
Query: 78 ASLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEG-NN 136
A YA R P + G + L + M + ++ W+ ++ YY+ +N
Sbjct: 223 AQRYANER------IPDCNLEHSMGPF-GENLAEGYGEMKGSDAVKFWLTEKPYYDHYSN 381
Query: 137 SCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
+CV + C YTQIVWR + +LGCA+A+C G +C Y PPGN+ GERP
Sbjct: 382 ACV--HDECLHYTQIVWRDSVHLGCARAKCN---NGWVFVICSYSPPGNIEGERP 531
>TC229998 weakly similar to PIR|T04232|T04232 pathogenesis-related protein
homolog F14M19.60 - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (78%)
Length = 915
Score = 66.6 bits (161), Expect = 7e-12
Identities = 48/152 (31%), Positives = 66/152 (42%), Gaps = 11/152 (7%)
Frame = +1
Query: 51 YLFEHNFVRREKG---------ISEHLEW-SDKLAKDASLYARFRRDKYLCTPGIFDKKR 100
+LF HN VR K + ++ W + + D L F D + I+
Sbjct: 199 FLFRHNLVRAAKWELPLMWDFQLEQYARWWAGERKADCKLEHSFPEDGFKLGENIY---- 366
Query: 101 YGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANYPCGDYTQIVWRKTKYL 159
W + P + +R W + YY N+CVP CG YTQIVW+ T+ +
Sbjct: 367 -------WGSGSA--WTPSDAVRAWADEEKYYTYATNTCVPGQM-CGHYTQIVWKSTRRI 516
Query: 160 GCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
GCA+ C D G V + C Y P GN GERP
Sbjct: 517 GCARVVC--DDGDVFM-TCNYDPVGNYVGERP 603
>TC227432 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (75%)
Length = 448
Score = 50.8 bits (120), Expect = 4e-07
Identities = 41/138 (29%), Positives = 64/138 (45%), Gaps = 1/138 (0%)
Frame = +2
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F+VL L L++V A A D ++ HN R + G+ ++ W D +A A YA
Sbjct: 59 FSVLCLLGLVIVGDHAAYAQ-DSPTDFVNAHNAARSQVGVP-NIVWDDTVAAFAQNYANQ 232
Query: 85 RRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPANY 143
R+ D K YG + L + + ++ WV ++ Y+ N N+CV
Sbjct: 233 RKGDCKLVHSGGDGK-YG----ENLAGSTGNLSGTNAVKLWVDEKSKYDYNSNTCVGGE- 394
Query: 144 PCGDYTQIVWRKTKYLGC 161
C YTQ+VW+ + LGC
Sbjct: 395 -CRHYTQVVWKNSVRLGC 445
>BG509716 similar to GP|4928711|gb|A PR1a precursor {Glycine max}, partial
(54%)
Length = 294
Score = 30.8 bits (68), Expect = 0.40
Identities = 22/58 (37%), Positives = 26/58 (43%), Gaps = 3/58 (5%)
Frame = +3
Query: 131 YYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQA---ECGPDYGGVRLNVCFYYPPGN 185
Y +NSCV C YTQ+VWR + G A+ GP G Y PPGN
Sbjct: 144 YDYNSNSCVGGE--CLHYTQVVWRDSVRXGXAKXAWENGGPXITG------NYAPPGN 293
>TC225700 similar to UP|V722_ARATH (P47192) Vesicle-associated membrane
protein 722 (AtVAMP722) (Synaptobrevin-related protein
1), complete
Length = 1191
Score = 27.7 bits (60), Expect = 3.4
Identities = 10/34 (29%), Positives = 19/34 (55%)
Frame = +3
Query: 132 YEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAE 165
+ +++C P G Y +I WRK+ GC++ +
Sbjct: 396 WTADSNCFP*TNQGGFYQEICWRKSSNSGCSEPQ 497
>TC212443
Length = 262
Score = 27.3 bits (59), Expect = 4.4
Identities = 13/35 (37%), Positives = 18/35 (51%)
Frame = +3
Query: 79 SLYARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTV 113
S Y F + +L + + R G CCN WL DT+
Sbjct: 138 SFYISFICEFFLFSSSL*LTVRSGSCCNFWLHDTL 242
>TC214232 weakly similar to UP|ATF5_HUMAN (Q9Y2D1) Cyclic-AMP-dependent
transcription factor ATF-5 (Activating transcription
factor 5) (Transcription factor ATFx), partial (8%)
Length = 1283
Score = 26.9 bits (58), Expect = 5.8
Identities = 14/47 (29%), Positives = 23/47 (48%)
Frame = +2
Query: 125 WVKDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYG 171
W ++R ++E + PC Y+ I R +K +G CGP+ G
Sbjct: 605 WTQERCFFEYHQKDCQVLMPC*HYSSIY*R*SK*VG-----CGPEVG 730
>CD395828 similar to PIR|T51584|T515 splicing factor 9G8-like SR protein 21
[validated] - Arabidopsis thaliana, partial (30%)
Length = 552
Score = 26.9 bits (58), Expect = 5.8
Identities = 14/30 (46%), Positives = 17/30 (56%)
Frame = +3
Query: 109 LEDTVKIMDPREVMRQWVKDRFYYEGNNSC 138
+E T DPREV V + F +EG NSC
Sbjct: 345 MEGTCCSCDPREVKCYVVLEIFSHEGYNSC 434
>TC225699 similar to UP|V722_ARATH (P47192) Vesicle-associated membrane
protein 722 (AtVAMP722) (Synaptobrevin-related protein
1), complete
Length = 1345
Score = 26.2 bits (56), Expect = 9.8
Identities = 9/31 (29%), Positives = 17/31 (54%)
Frame = +3
Query: 132 YEGNNSCVPANYPCGDYTQIVWRKTKYLGCA 162
+ +++C P G Y ++ WRK+ GC+
Sbjct: 411 WTADSNCFPRTNQGGFYQEVCWRKSCNSGCS 503
>TC203702 weakly similar to GB|AAP68215.1|31711718|BT008776 At1g51580
{Arabidopsis thaliana;} , partial (25%)
Length = 1642
Score = 26.2 bits (56), Expect = 9.8
Identities = 14/39 (35%), Positives = 17/39 (42%)
Frame = +2
Query: 137 SCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRL 175
SC + +PC T W TK C A C GG+ L
Sbjct: 86 SCTRSCHPCSLSTYRDWI*TKCCSCC*ASCAFTTGGLPL 202
>TC204744 homologue to UP|Q9MB06 (Q9MB06) Type 2A protein phosphatase-1,
complete
Length = 1462
Score = 26.2 bits (56), Expect = 9.8
Identities = 20/59 (33%), Positives = 27/59 (44%), Gaps = 6/59 (10%)
Frame = +1
Query: 17 PKMFPHPFFAVLVALALLLVSSQAT----PATTD--GAKVYLFEHNFVRREKGISEHLE 69
PK F PFF L + LLL+ S + P T D GA E N +R + + H +
Sbjct: 76 PKRFVLPFFGTLPSAPLLLLRSPSAILR*PNTIDPSGASDLGSERNESKRNETMPSHAD 252
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,319,593
Number of Sequences: 63676
Number of extensions: 183744
Number of successful extensions: 909
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 894
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 897
length of query: 191
length of database: 12,639,632
effective HSP length: 92
effective length of query: 99
effective length of database: 6,781,440
effective search space: 671362560
effective search space used: 671362560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0323a.9


